8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8BZ1
 
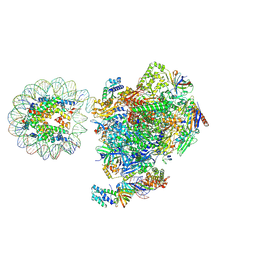 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8BVW
 
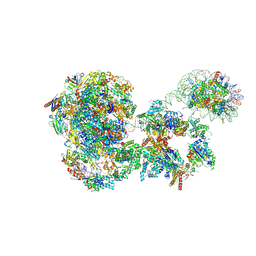 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
9GD0
 
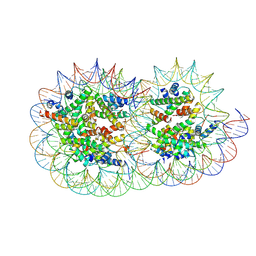 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
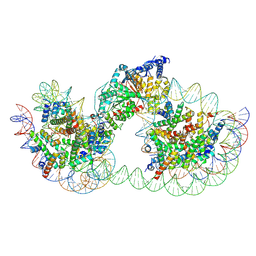 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD3
 
 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
4BXZ
 
 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BY7
 
 | | elongating RNA Polymerase II-Bye1 TLD complex | | Descriptor: | , 5'-D(*DAP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*DTP)-3', 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
4BY1
 
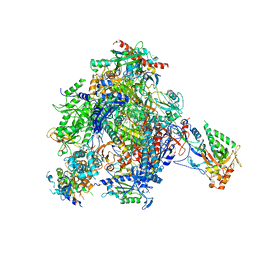 | | elongating RNA Polymerase II-Bye1 TLD complex soaked with AMPCPP | | Descriptor: | 5'-D(*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP *TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*AP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BXX
 
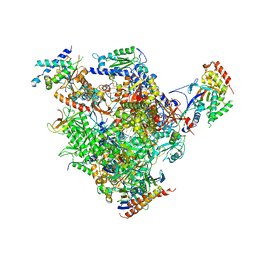 | | Arrested RNA polymerase II-Bye1 complex | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*AP*CP*CP*TP*GP *GP*TP*GP* BRUP*TP*GP*CP*TP*CP*TP*AP*AP*DC)-3', 5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP)-3', 5'-D(*GP*AP*GP*GP*TP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9I81
 
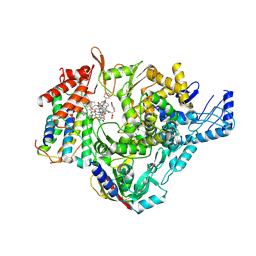 | | SARS-CoV-2 RdRp bound to a stack of three HeE1-2Tyr molecules | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kabinger, F, Doze, V, Schmitzova, J, Lidschreiber, M, Dienemann, C, Cramer, P. | | Deposit date: | 2025-02-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of SARS-CoV-2 polymerase inhibition by nonnucleoside inhibitor HeE1-2Tyr.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
2MOW
 
 | |
7A9W
 
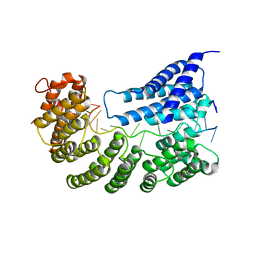 | | Structure of yeast Rmd9p in complex with 20nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A9X
 
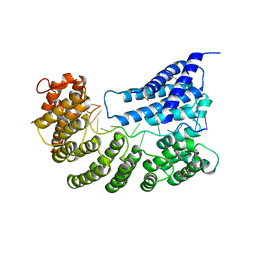 | | Structure of yeast Rmd9p in complex with 16nt target RNA | | Descriptor: | CHLORIDE ION, Protein RMD9, mitochondrial, ... | | Authors: | Hillen, H.S, Markov, D.A, Ireneusz, W.D, Hofmann, K.B, Cowan, A.T, Jones, J.L, Temiakov, D, Cramer, P, Anikin, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The pentatricopeptide repeat protein Rmd9 recognizes the dodecameric element in the 3'-UTRs of yeast mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8RAO
 
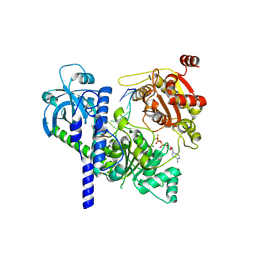 | | Structure of Sen1-ADP.BeF3-RNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Helicase SEN1, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8RAP
 
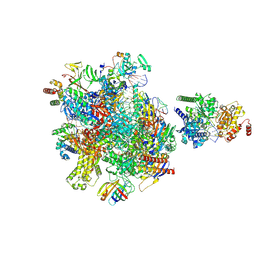 | | Structure of Sen1-ADP.BeF3 bound RNA Polymerase II pre-termination complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8RAM
 
 | | Structure of Sen1 bound RNA Polymerase II pre-termination complex | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Rengachari, S, Lidscreiber, M, Cramer, P. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of polyadenylation-independent RNA polymerase II termination.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8RAN
 
 | |
