7O7C
 
 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7U
 
 | | Crystal structure of rsEGFP2 in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Woodhouse, J, Coquelle, N, Barends, T.R.M, Schlichting, I, Weik, M, Colletier, J.-P. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5HD3
 
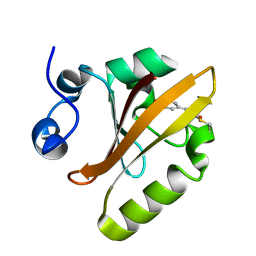 | |
5HDC
 
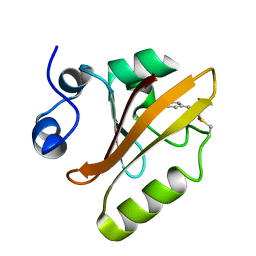 | |
5HDD
 
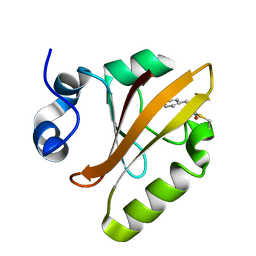 | |
5HDS
 
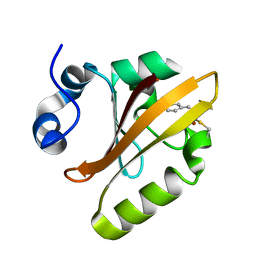 | |
5HD5
 
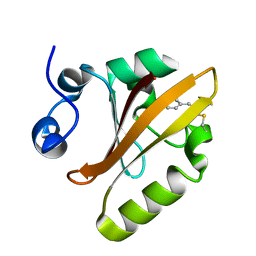 | |
5MND
 
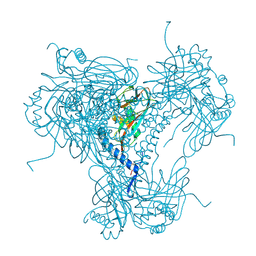 | |
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NM4
 
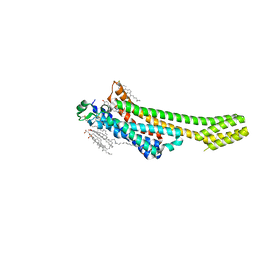 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NQU
 
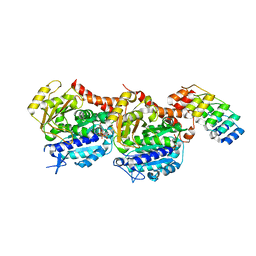 | | Tubulin Darpin cryo structure | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NM5
 
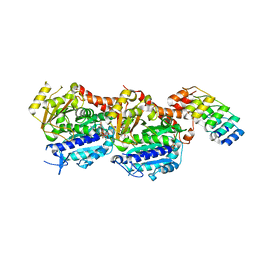 | | Tubulin Darpin room-temperature structure in complex with Colchicine determined by serial millisecond crystallography | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NJM
 
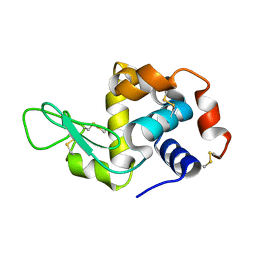 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NQT
 
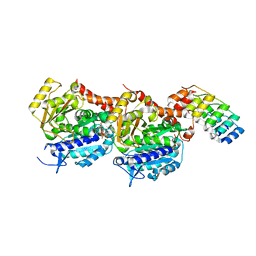 | | Tubulin Darpin room-temperature structure determined by serial millisecond crystallography | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5O5W
 
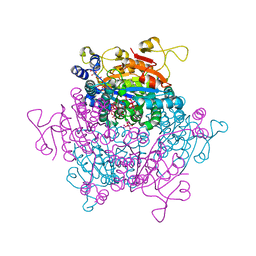 | | Molybdenum storage protein room-temperature structure determined by serial millisecond crystallography | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steffen, B, Weinert, T, Ermler, U, Standfuss, J. | | Deposit date: | 2017-06-02 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5LBR
 
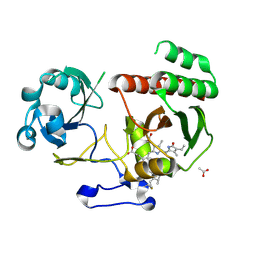 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at SACLA | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | Authors: | Edlund, P, Claesson, E, Nakane, T, Takala, H, Dods, R, Schmidt, M, Westenhoff, S. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
5L8M
 
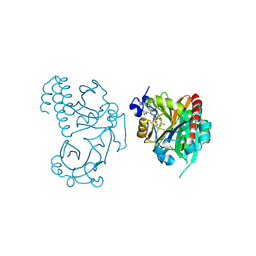 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at LCLS | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Edlund, P, Henry, L, Dods, R, Schmidt, M, Westenhoff, S. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
5K5B
 
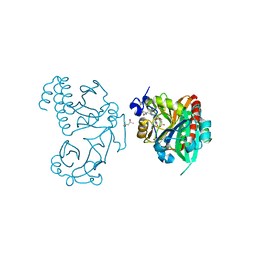 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans BphP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Takala, H, Edlund, P, Claesson, E, Ihalainen, J.A, Westenhoff, S. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
5TRX
 
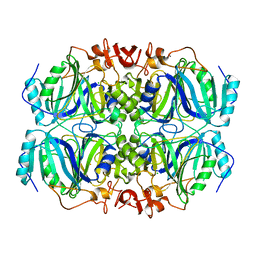 | | Room temperature structure of an extradiol ring-cleaving dioxygenase from B.fuscum determined using serial femtosecond crystallography | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Oberthuer, D, Tolstikova, A, Mariani, V. | | Deposit date: | 2016-10-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography
Sci. Rep., 7, 2017
|
|
5U5Q
 
 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
