1DSD
 
 | |
1DSC
 
 | |
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
6YP7
 
 | | PSII-LHCII C2S2 supercomplex from Pisum sativum grown in high light conditions | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Grinzato, A, Albanese, P, Zanotti, G, Pagliano, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants.
Int J Mol Sci, 21, 2020
|
|
3F3H
 
 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
8GNA
 
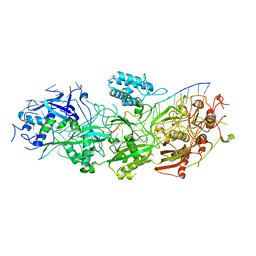 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8GU6
 
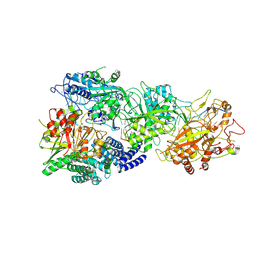 | | Structure of the SbCas7-11-crRNA-NTR-Csx29 complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-09-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8HF2
 
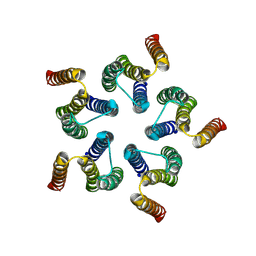 | | Cryo-EM structure of WeiTsing | | Descriptor: | PRA1 family protein | | Authors: | Qin, L, Tang, L.H, Chen, Y.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance.
Cell, 186, 2023
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
7YTL
 
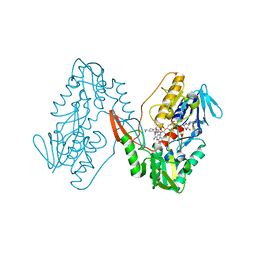 | | Structure of a oxidoreductase in complex with quinone | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE, UBIQUINONE-1 | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
8AWW
 
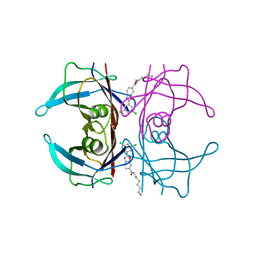 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
9EOU
 
 | |
6AZ0
 
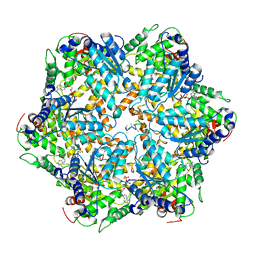 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
5NEW
 
 | |
5CR4
 
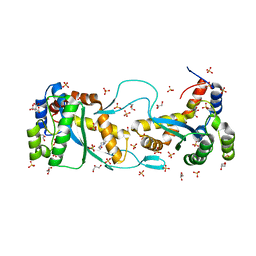 | |
2XWY
 
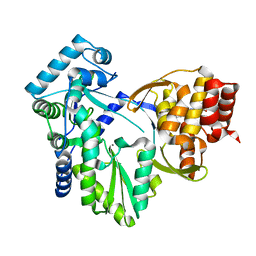 | | Structure of MK-3281, a Potent Non-Nucleoside Finger-Loop Inhibitor, in complex with the Hepatitis C Virus NS5B Polymerase | | Descriptor: | (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl)amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Di Marco, S, Baiocco, P. | | Deposit date: | 2010-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of (7R)-14-Cyclohexyl-7-{[2-(Dimethylamino)Ethyl] (Methyl)Amino}-7,8-Dihydro-6H-Indolo[1,2-E][1,5] Benzoxazocine -11-Carboxylic Acid (Mk-3281), a Potent and Orally Bioavailable Finger-Loop Inhibitor of the Hepatitis C Virus Ns5B Polymerase
J.Med.Chem., 54, 2011
|
|
3EBN
 
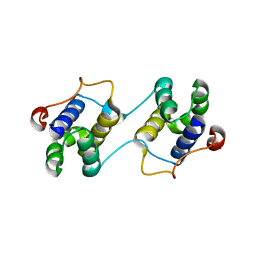 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | Deposit date: | 2008-08-28 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
6FI8
 
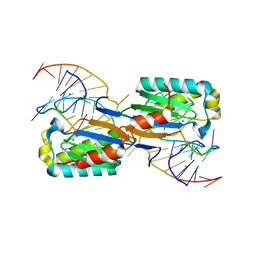 | |
6ITE
 
 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
2KLQ
 
 | | The solution structure of CBD of human MCM6 | | Descriptor: | DNA replication licensing factor MCM6 | | Authors: | Liu, C. | | Deposit date: | 2009-07-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6
J.Biol.Chem., 285, 2010
|
|
2LE8
 
 | | The protein complex for DNA replication | | Descriptor: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | Authors: | Liu, C, Wei, Z, Zhu, G. | | Deposit date: | 2011-06-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
2K7X
 
 | |
2XA0
 
 | | Crystal structure of BCL-2 in complex with a BAX BH3 peptide | | Descriptor: | APOPTOSIS REGULATOR BAX, APOPTOSIS REGULATOR BCL-2 | | Authors: | Ku, B, Oh, B.H. | | Deposit date: | 2010-03-25 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evidence that Inhibition of Bax Activation by Bcl- 2 Involves its Tight and Preferential Interaction with the Bh3 Domain of Bax.
Cell Res., 21, 2011
|
|
3BL2
 
 | | Crystal Structure of M11, the BCL-2 Homolog of Murine Gamma-herpesvirus 68, Complexed with Mouse Beclin1 (residues 106-124) | | Descriptor: | Beclin-1, V-bcl-2 | | Authors: | Oh, B.-H, Woo, J.-S, Ku, B. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine gamma-Herpesvirus 68
Plos Pathog., 4, 2008
|
|
