2R2V
 
 | |
6INH
 
 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6INF
 
 | | a glycosyltransferase complex with UDP | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, X, Yang, T, Naismith, J.H. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6ING
 
 | |
2YHV
 
 | |
2YFX
 
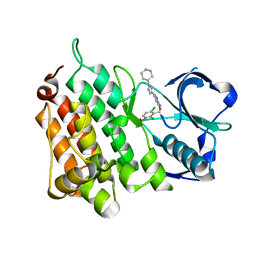 | | Structure of L1196M Mutant Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A. | | Deposit date: | 2011-04-08 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Potent and Selective Inhibitors to Overcome Clinical Anaplastic Lymphoma Kinase Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4ANQ
 
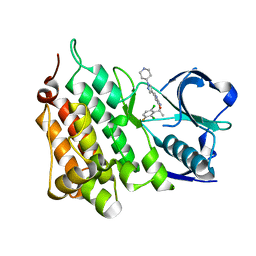 | | Structure of G1269A Mutant Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of Potent and Selective Inhibitors to Overcome Clinical Anaplastic Lymphoma Kinase Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4ANL
 
 | |
4CCB
 
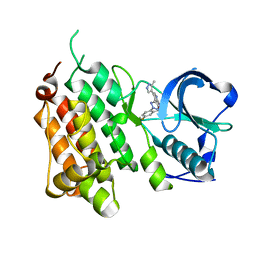 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 3-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)-5-(5-methyl-1H- pyrazol-4-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-[5-fluoranyl-2-(1,2,3-triazol-2-yl)phenyl]ethoxy]-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CCU
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy) pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol | | Descriptor: | 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CD0
 
 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2- yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propane-1,2-diol | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4Z8I
 
 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
4ZXM
 
 | | Crystal structure of PGRP domain from Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | PGRP domain of peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Yu, H.M, Luo, M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
6XVF
 
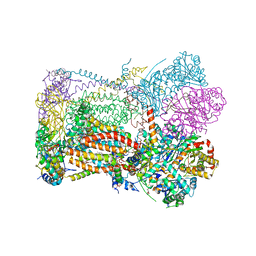 | | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites.
Front Cell Infect Microbiol, 10, 2020
|
|
8BBH
 
 | |
4U9A
 
 | |
4YVV
 
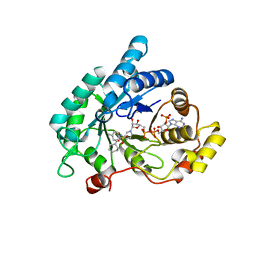 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
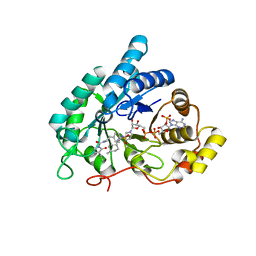 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVP
 
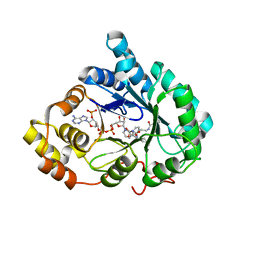 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4ZFC
 
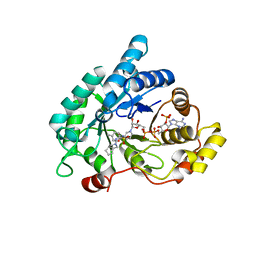 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4U97
 
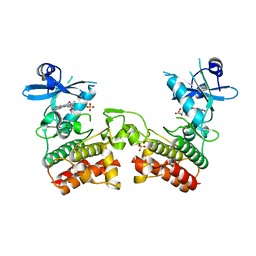 | | Crystal Structure of Asymmetric IRAK4 Dimer | | Descriptor: | Interleukin-1 receptor-associated kinase 4, STAUROSPORINE, SULFATE ION | | Authors: | Ferrao, R, Wu, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | IRAK4 Dimerization and trans-Autophosphorylation Are Induced by Myddosome Assembly.
Mol.Cell, 55, 2014
|
|
6IH5
 
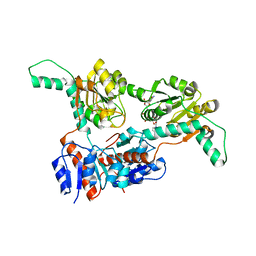 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6IH2
 
 | |
6IJ1
 
 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
