7XVV
 
 | |
7XVW
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVU
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YK2
 
 | | Cryo-EM structure of Apo-alpha-syn fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xu, Q.H, Xia, W.C, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Dynamics of an alpha-Synuclein Fibril upon Receptor Binding Revealed by Insensitive Nuclei Enhanced by Polarization Transfer-Based Solid-State Nuclear Magnetic Resonance and Cryo-Electron Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7YK8
 
 | | Cryo-EM structure of dLAG3-alpha-syn fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xu, Q.H, Xia, W.C, Tao, Y.Q, Liu, C. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational Dynamics of an alpha-Synuclein Fibril upon Receptor Binding Revealed by Insensitive Nuclei Enhanced by Polarization Transfer-Based Solid-State Nuclear Magnetic Resonance and Cryo-Electron Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
6IIH
 
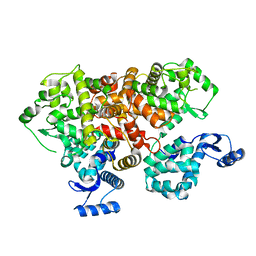 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
5GJ6
 
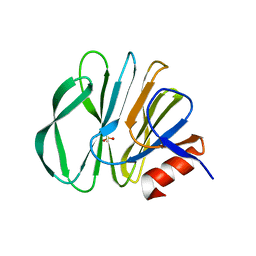 | |
8H05
 
 | |
8H04
 
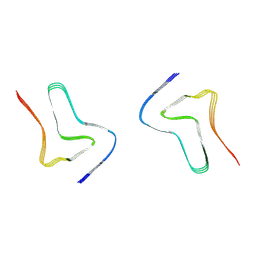 | |
7W26
 
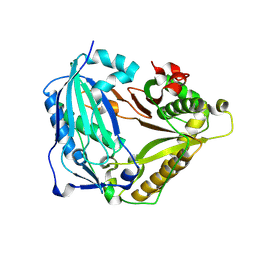 | | monolignol ferulate transferase | | Descriptor: | Ferulate monolignol transferase | | Authors: | Xi, L, Shuliu, D, Yue, F, Yi, Z. | | Deposit date: | 2021-11-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of the plant feruloyl-coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates.
Biochem.Biophys.Res.Commun., 594, 2022
|
|
7KLM
 
 | | Human Arginase1 Complexed with Inhibitor Compound 24a | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLL
 
 | | Human Arginase1 Complexed with Inhibitor Compound 18 | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-azanyl-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLK
 
 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7JTP
 
 | | Crystal structure of Protac MS67 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | Elongin-B, Elongin-C, GLYCEROL, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
7JTO
 
 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LWJ
 
 | |
7LWI
 
 | |
7LWM
 
 | |
7LWW
 
 | |
7LWO
 
 | |
7LYM
 
 | |
7LWT
 
 | |
7LWU
 
 | |
7LWV
 
 | |
