1Y0H
 
 | | Structure of Rv0793 from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, hypothetical protein Rv0793 | | Authors: | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
2NR9
 
 | | Crystal structure of GlpG, Rhomboid Peptidase from Haemophilus influenzae | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, Protein glpG homolog | | Authors: | Lemieux, M.J, Fischer, S.J, Cherney, M.M, Bateman, K.S, James, M.N.G. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2GK1
 
 | | X-ray crystal structure of NGT-bound HexA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit alpha, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystallographic Structure of Human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of G(M2) Ganglioside Hydrolysis.
J.Mol.Biol., 359, 2006
|
|
2GJX
 
 | | Crystallographic structure of human beta-Hexosaminidase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase alpha chain, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic structure of human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss
of GM2 Ganglioside Hydrolysis
J.Mol.Biol., 359, 2006
|
|
7DMX
 
 | |
7E9Y
 
 | | Crystal structure of eLACCO1 | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CALCIUM ION, Lactate-binding periplasmic protein TTHA0766,Lactate-binding periplasmic protein TTHA0766 | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J, Nasu, Y. | | Deposit date: | 2021-03-05 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A genetically encoded fluorescent biosensor for extracellular L-lactate.
Nat Commun, 12, 2021
|
|
9CR1
 
 | | Crystal structure of histidine racemase (HisR) of Fusobacterium nucleatum (C67S) | | Descriptor: | Histidine racemase, SULFATE ION | | Authors: | Chen, P, Lamer, T, Vederas, J.C, Lemieux, M.J. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, characterization, and structure of a cofactor-independent histidine racemase from the oral pathogen Fusobacterium nucleatum.
J.Biol.Chem., 300, 2024
|
|
9CR6
 
 | | Crystal structure of histidine racemase (HisR) of Fusobacterium nucleatum (C209S) | | Descriptor: | Histidine racemase, PHOSPHATE ION | | Authors: | Chen, P, Lamer, T, Vederas, J.C, Lemieux, M.J. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery, characterization, and structure of a cofactor-independent histidine racemase from the oral pathogen Fusobacterium nucleatum.
J.Biol.Chem., 300, 2024
|
|
4U9C
 
 | |
6WTJ
 
 | | Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 11, 2020
|
|
9BPF
 
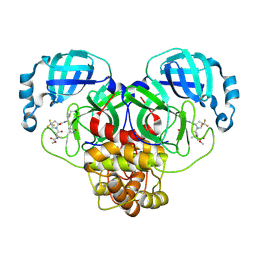 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BOO
 
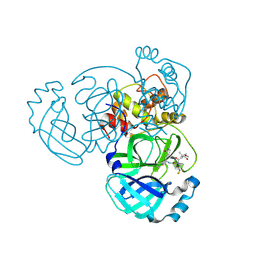 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
7R7H
 
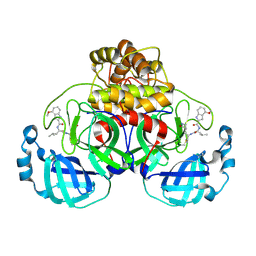 | | Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors.
Rsc Med Chem, 12, 2021
|
|
3ODJ
 
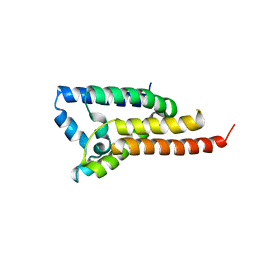 | | Crystal structure of H. influenzae rhomboid GlpG with disordered loop 4, helix 5 and loop 5 | | Descriptor: | Rhomboid protease glpG | | Authors: | Brooks, C.L, Lazareno-Saez, C, Lamoureux, J.S, Mak, M.W, Lemieux, M.J. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Insights into Substrate Gating in H. influenzae Rhomboid.
J.Mol.Biol., 407, 2011
|
|
7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
9EJN
 
 | | Crystal structure of magnesium-transporting ATPase MgtA in an E1-like magnesium-bound state | | Descriptor: | MAGNESIUM ION, Magnesium-transporting ATPase, P-type 1 | | Authors: | Khan, M.B, Primeau, J.O, Basu, P.C, Morth, J.P, Lemieux, M.J, Young, H.S. | | Deposit date: | 2024-11-28 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Crystal structure of magnesium-transporting ATPase MgtA in an E1-like magnesium-bound state
To Be Published
|
|
3UAQ
 
 | | Crystal Structure of the N-lobe Domain of Lactoferrin Binding Protein B (LbpB) of Moraxella bovis | | Descriptor: | LbpB B-lobe | | Authors: | Arutyunova, E, Brooks, C.L, Beddeck, A, Mak, M.W, Schryvers, A.B, Lemieux, M.J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9318 Å) | | Cite: | Crystal structure of the N-lobe of lactoferrin binding protein B from Moraxella bovis(1).
Biochem.Cell Biol., 90, 2012
|
|
7YV3
 
 | |
7YV5
 
 | |
7SMV
 
 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
7SNA
 
 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
6WTK
 
 | | Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 11, 2020
|
|
6WTM
 
 | |
4HDD
 
 | | Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease | | Descriptor: | ACETATE ION, Rhomboid protease GlpG | | Authors: | Lazareno-Saez, C, Arutyunova, E, Coquelle, N, Lemieux, M.J. | | Deposit date: | 2012-10-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease.
J.Mol.Biol., 425, 2013
|
|
1PW4
 
 | | Crystal Structure of the Glycerol-3-Phosphate Transporter from E.Coli | | Descriptor: | Glycerol-3-phosphate transporter | | Authors: | Huang, Y, Lemieux, M.J, Song, J, Auer, M, Wang, D.N. | | Deposit date: | 2003-06-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia Coli
Science, 301, 2003
|
|
