6R5R
 
 | |
3TD5
 
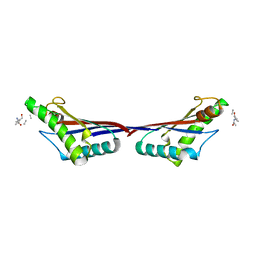 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
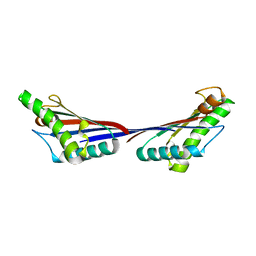 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
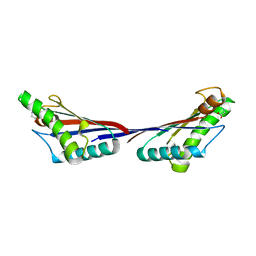 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
7OK9
 
 | |
7O4A
 
 | |
7O49
 
 | |
7O4C
 
 | |
7O4B
 
 | |
3KQI
 
 | | crystal structure of PHF2 PHD domain complexed with H3K4Me3 peptide | | Descriptor: | CHLORIDE ION, GLYCEROL, H3K4Me3 peptide, ... | | Authors: | Wen, H, Li, J.Z, Song, T, Lu, M, Lee, M. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation.
J.Biol.Chem., 285, 2010
|
|
4PB2
 
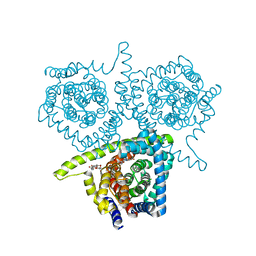 | | Structure of vcCNT-7C8C bound to 5-fluorouridine | | Descriptor: | 5-FLUOROURIDINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD7
 
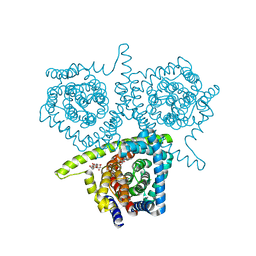 | | Structure of vcCNT bound to zebularine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD6
 
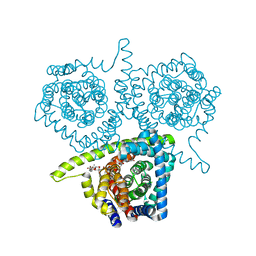 | | Crystal structure of vcCNT-7C8C bound to uridine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD8
 
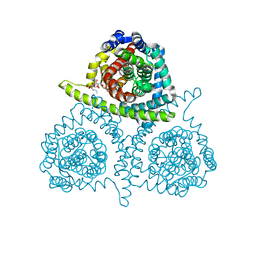 | | Structure of vcCNT-7C8C bound to pyrrolo-cytidine | | Descriptor: | 6-methyl-3-(beta-D-ribofuranosyl)-3,7-dihydro-2H-pyrrolo[2,3-d]pyrimidin-2-one, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD5
 
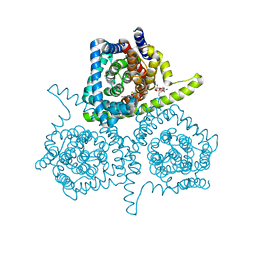 | | Crystal structure of vcCNT-7C8C bound to gemcitabine | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, GEMCITABINE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PDA
 
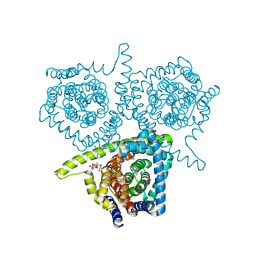 | | Structure of vcCNT-7C8C bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PB1
 
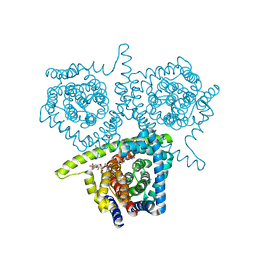 | | Structure of vcCNT-7C8C bound to ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD9
 
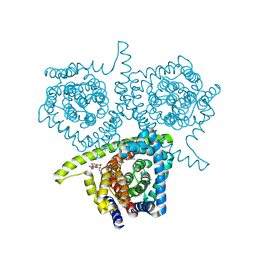 | | Structure of vcCNT-7C8C bound to adenosine | | Descriptor: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
8C54
 
 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
1C29
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-23 | | Release date: | 2000-01-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1C9D
 
 | |
1C8V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
8DA2
 
 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
8ELI
 
 | |
8EUW
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
