8E4F
 
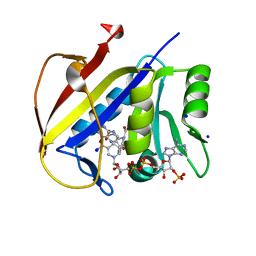 | | Crystal structure of dihydrofolate reductase (DHFR) from the filarial nematode W. bancrofti in complex with NADPH and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lange, K, Frey, K.M, Goodey, N.M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of dihydrofolate reductase from the filarial nematode W. bancrofti in complex with NADPH and folate.
Plos Negl Trop Dis, 17, 2023
|
|
2Y8G
 
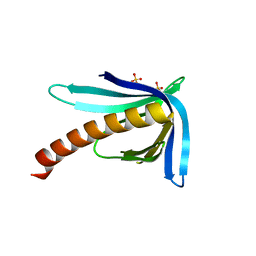 | | Structure of the Ran-binding domain from human RanBP3 (E352A-R353V double mutant) | | Descriptor: | RAN-BINDING PROTEIN 3, SULFATE ION | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
2Y8F
 
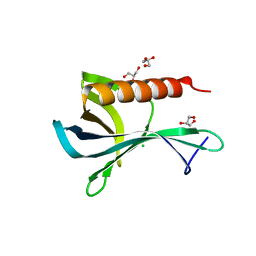 | | Structure of the Ran-binding domain from human RanBP3 (wild type) | | Descriptor: | CHLORIDE ION, GLYCEROL, RAN-BINDING PROTEIN 3 | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
1WQJ
 
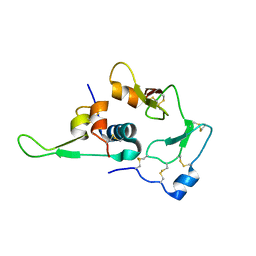 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
1BOE
 
 | | STRUCTURE OF THE IGF BINDING DOMAIN OF THE INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5): IMPLICATIONS FOR IGF AND IGF-I RECEPTOR INTERACTIONS | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5)) | | Authors: | Kalus, W, Zweckstetter, M, Renner, C, Sanchez, Y, Georgescu, J, Grol, M, Demuth, D, Schumacherdony, C, Lang, K, Holak, T.H. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions.
EMBO J., 17, 1998
|
|
3C3Z
 
 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to ribostamycin | | Descriptor: | HIV-1 subtype F genomic RNA, RIBOSTAMYCIN | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3C7R
 
 | | Crystal Structure of HIV-1 subtype F DIS extended duplex RNA bound to neomycin | | Descriptor: | HIV-1 subtype F genomic RNA, NEOMYCIN, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3C5D
 
 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to lividomycin | | Descriptor: | 'HIV-1 subtype F genomic RNA, (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DVZ
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW7
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
5O74
 
 | | Crystal structure of human Rab1b covalently bound to the GEF domain of DrrA/SidM from Legionella pneumophila in the presence of GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Multifunctional virulence effector protein DrrA, Ras-related protein Rab-1B | | Authors: | Cigler, M, Mueller, T, Horn-Ghetko, D, von Wrisberg, M.K, Fottner, M, Goody, R.S, Itzen, A, Mueller, M.P, Lang, K. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proximity-Triggered Covalent Stabilization of Low-Affinity Protein Complexes In Vitro and In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4AQY
 
 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
6YX5
 
 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
9CS7
 
 | | Structure of Azospirillum bacterial CARD crystal form 1 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, HIll, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
9CS8
 
 | | Structure of Azospirillum bacterial CARD crystal form 2 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, Hill, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
4NHD
 
 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
