2L99
 
 | | Solution structure of LAK160-P10 | | Descriptor: | LAK160-P10 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L9A
 
 | | Solution structure of LAK160-P12 | | Descriptor: | LAK160-P12 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L96
 
 | | Solution structure of LAK160-P7 | | Descriptor: | LAK160-P7 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
6NZK
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2FJN
 
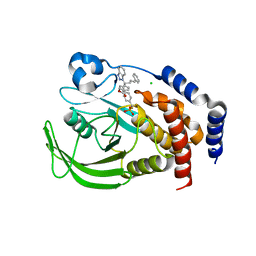 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, Tyrosine-protein phosphatase, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Kennedy, B.P, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
2FJM
 
 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
6QFY
 
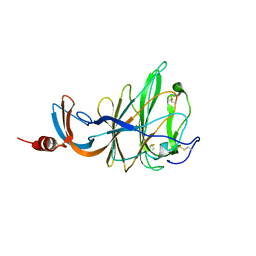 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6MWY
 
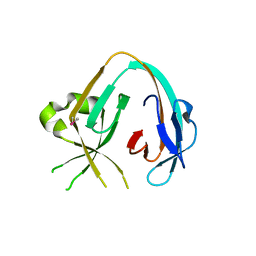 | | The Prp8 intein of Cryptococcus gattii | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Fu, B, Green, C.M, Lang, Y, Zhang, J, Oven, T.S, Li, X, Callahan, B.P, Chaturvedi, S, Belfort, M, Liao, G, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
4LR6
 
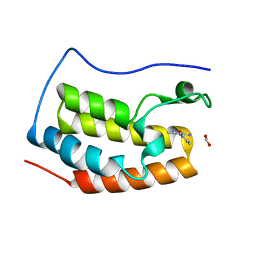 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
1QDD
 
 | | CRYSTAL STRUCTURE OF HUMAN LITHOSTATHINE TO 1.3 A RESOLUTION | | Descriptor: | LITHOSTATHINE, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Gerbaud, V, Pignol, D, Loret, E, Bertrand, J.A, Berland, Y, Fontecilla-Camps, J.C, Canselier, J.P, Gabas, N, Verdier, J.M. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of calcite crystal growth inhibition by the N-terminal undecapeptide of lithostathine.
J.Biol.Chem., 275, 2000
|
|
5KR7
 
 | | KDM4C bound to pyrazolo-pyrimidine scaffold | | Descriptor: | 6-ethyl-2,5-dimethyl-7-oxidanylidene-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bellon, S.F, Poy, F, Setser, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent, selective KDM5 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6WMI
 
 | | ZNF410 zinc fingers 1-5 with 17 mer blunt DNA Oligonucleotide | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*CP*AP*TP*AP*AP*TP*AP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*TP*AP*TP*GP*GP*GP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Ren, R, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression.
Mol.Cell, 81, 2021
|
|
7RIO
 
 | | human PXR LBD bound to GSK003 | | Descriptor: | Isoform 1C of Nuclear receptor subfamily 1 group I member 2, N-[3-(2-tert-butyl-5-{2-[2-(methanesulfonyl)ethyl]pyrimidin-4-yl}-1,3-thiazol-4-yl)-2-fluorophenyl]-2,5-difluorobenzene-1-sulfonamide | | Authors: | Williams, S.P, Wisely, G.B, Ward, P. | | Deposit date: | 2021-07-20 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7RIV
 
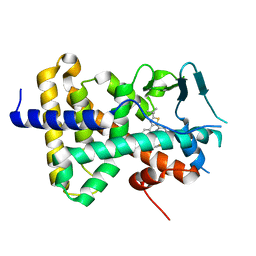 | | human PXR LBD bound to GSK001 | | Descriptor: | Dabrafenib, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ward, P. | | Deposit date: | 2021-07-20 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7RIU
 
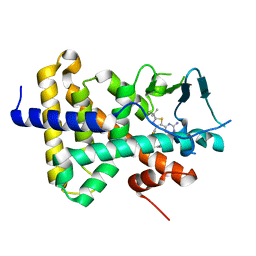 | | human PXR LBD bound to GSK002 | | Descriptor: | Isoform 1C of Nuclear receptor subfamily 1 group I member 2, N-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}thiophene-3-sulfonamide | | Authors: | Williams, S.P, Wisely, G.B, Ward, P. | | Deposit date: | 2021-07-20 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7N2A
 
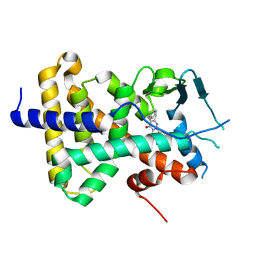 | | human PXR LBD bound to compound 2 | | Descriptor: | 5-benzyl-2-(3-fluoro-2-hydroxyphenyl)-6-methyl-3-(2-phenylethyl)pyrimidin-4(3H)-one, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ramanjulu, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
7XZ5
 
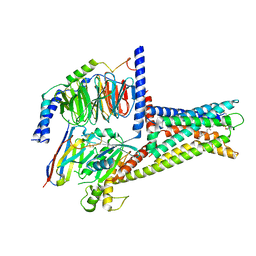 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XZ6
 
 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
6Y3Y
 
 | | Human Coronavirus HKU1 Haemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Hurdiss, D.L, Drulyte, I, Pronker, M.F. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans.
Nat Commun, 11, 2020
|
|
4X2I
 
 | | Discovery of benzotriazolo diazepines as orally-active inhibitors of BET bromodomains: Crystal structure of BRD4 with CPI-13 | | Descriptor: | (4R)-6-(4-chlorophenyl)-1,4-dimethyl-5,6-dihydro-4H-[1,2,4]triazolo[4,3-a][1,5]benzodiazepine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Bellon, S.F, Jayaram, H, Poy, F. | | Deposit date: | 2014-11-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Benzotriazolo[4,3-d][1,4]diazepines as Orally Active Inhibitors of BET Bromodomains.
Acs Med.Chem.Lett., 7, 2016
|
|
3CWE
 
 | | PTP1B in complex with a phosphonic acid inhibitor | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1, [{2-bromo-4-[(2R)-3-oxo-2,3-diphenylpropyl]phenyl}(difluoro)methyl]phosphonic acid | | Authors: | Scapin, G, Han, Y, Kennedy, B.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
