8JYY
 
 | |
7WN8
 
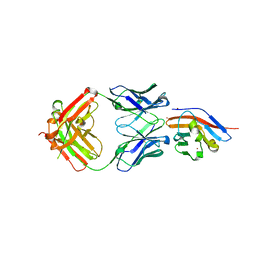 | | Crystal structure of antibody (BC31M5) binds to CD47 | | Descriptor: | BC31M5 Fab Heavy chain, BC31M5 Fab Light chain, Leukocyte surface antigen CD47, ... | | Authors: | Li, Y, Wang, W, Sui, J, Zhang, S. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A pH-dependent anti-CD47 antibody that selectively targets solid tumors and improves therapeutic efficacy and safety.
J Hematol Oncol, 16, 2023
|
|
6N1H
 
 | | Cryo-EM structure of ASC-CARD filament | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6O59
 
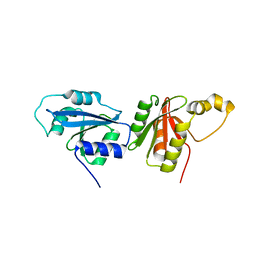 | |
8GJZ
 
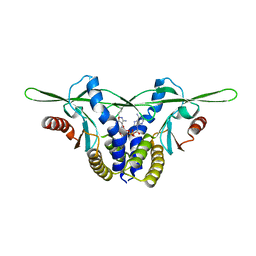 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
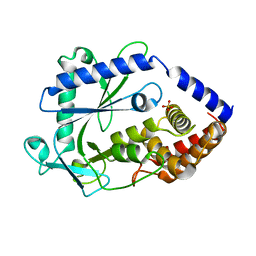 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
5YDM
 
 | |
5YDL
 
 | |
5YDA
 
 | |
7CWW
 
 | | Crystal structure of TsrL | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, TsrE | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-08-31 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of TsrL
To Be Published
|
|
7UR2
 
 | |
7F68
 
 | | Crystal structure of N-ras S89D | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, Y, Sun, Q. | | Deposit date: | 2021-06-24 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of N-ras S89D
To Be Published
|
|
7YTD
 
 | | Cryo-EM structure of four human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YSG
 
 | | Cryo-EM structure of human FcmR bound to sIgM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTC
 
 | | Cryo-EM structure of human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTE
 
 | |
8XAB
 
 | |
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
4WSI
 
 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7P8A
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_003 | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N},9-dimethylpurin-6-amine | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8B
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_006 | | Descriptor: | 9-cyclopropyl-~{N}-methyl-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P87
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_001 | | Descriptor: | N-methyl-4,5,6,7-tetrahydro-1H-indazole-3-carboxamide, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P88
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_002 | | Descriptor: | 2-chloranyl-~{N}-methyl-9~{H}-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
