1VET
 
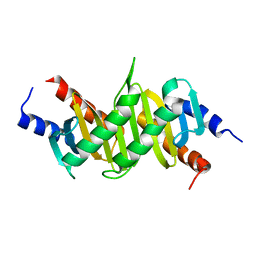 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VEU
 
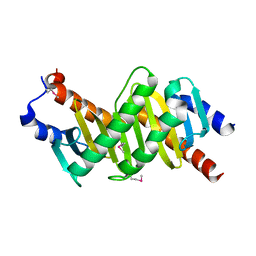 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SOT
 
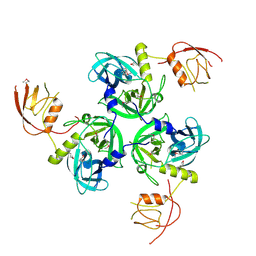 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1SOZ
 
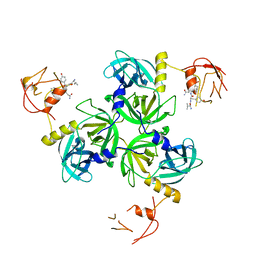 | | Crystal Structure of DegS protease in complex with an activating peptide | | Descriptor: | Protease degS, activating peptide | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
1VCW
 
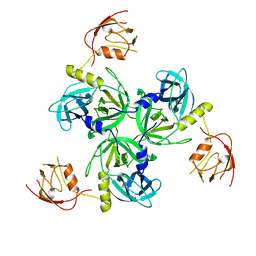 | | Crystal structure of DegS after backsoaking the activating peptide | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease.
Cell(Cambridge,Mass.), 117, 2004
|
|
4C2D
 
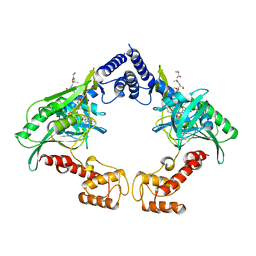 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2E
 
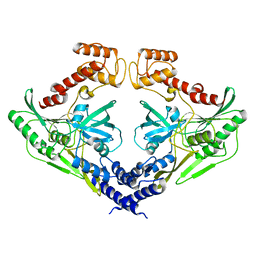 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2C
 
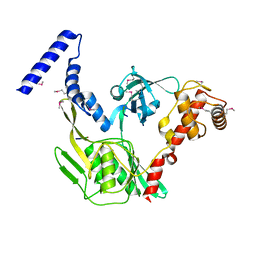 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2G
 
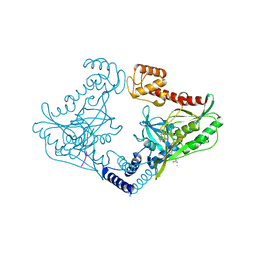 | | Crystal structure of CtpB(S309A) in complex with a peptide having a Val-Pro-Ala C-terminus | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2H
 
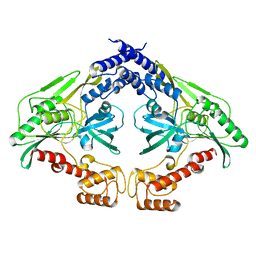 | | Crystal structure of the CtpB(V118Y) mutant | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2F
 
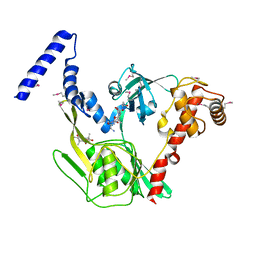 | | Crystal structure of the CtpB R168A mutant present in an active conformation | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
6QDJ
 
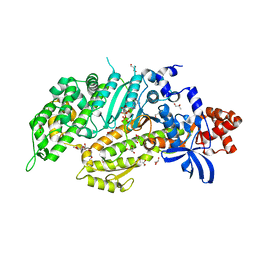 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
5HBN
 
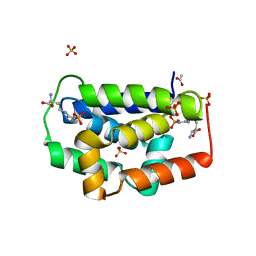 | |
7BII
 
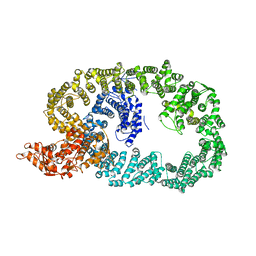 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
6TV6
 
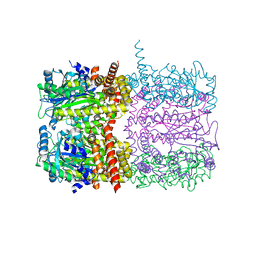 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
5D4W
 
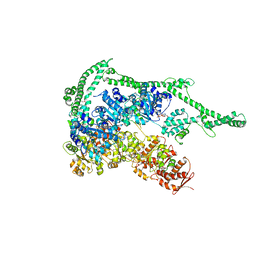 | | Crystal structure of Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative heat shock protein | | Authors: | Heuck, A, Schitter-Sollner, S, Clausen, T. | | Deposit date: | 2015-08-09 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the disaggregase activity and regulation of Hsp104.
Elife, 5, 2016
|
|
2R3Y
 
 | |
6QDL
 
 | |
6QDM
 
 | |
6QDK
 
 | |
7AA4
 
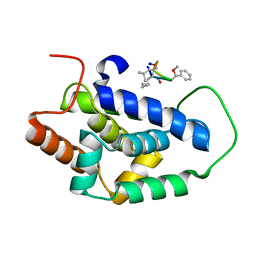 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
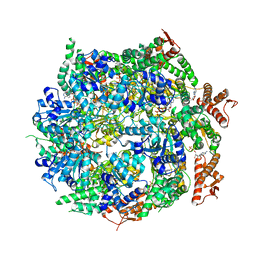 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
8ATX
 
 | |
8AUW
 
 | |
