5ZXA
 
 | | Crystal structure of fibronectin-binding protein Apa mutant from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL, MERCURY (II) ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5ZX9
 
 | | Crystal structure of apo form fibronectin-binding protein Apa from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5UN6
 
 | |
5UN5
 
 | |
5GM4
 
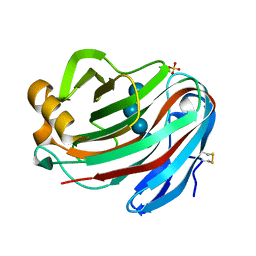 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM3
 
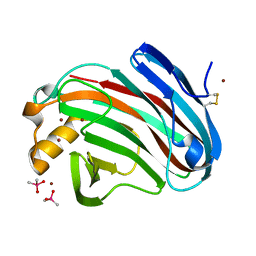 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 | | Descriptor: | CACODYLATE ION, Endoglucanase-1, ZINC ION | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM9
 
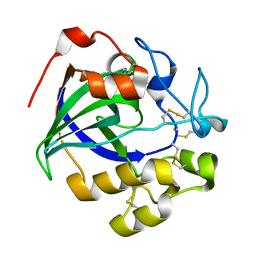 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GLY
 
 | | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GLX
 
 | | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GM5
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoglucanase-1, SULFATE ION, ... | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
2Z9L
 
 | |
2ZU3
 
 | | Complex structure of CVB3 3C protease with TG-0204998 | | Descriptor: | 3C proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Lee, C.C, Tsui, Y.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZTZ
 
 | |
2Z94
 
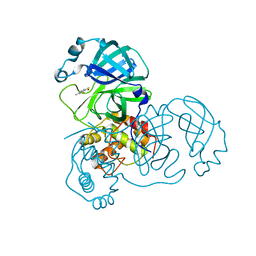 | | Complex structure of SARS-CoV 3C-like protease with TDT | | Descriptor: | 4-methylbenzene-1,2-dithiol, Replicase polyprotein 1ab, ZINC ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-17 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors
Febs Lett., 581, 2007
|
|
2Z9K
 
 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZU2
 
 | | complex structure of CoV 229E 3CL protease with EPDTC | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTY
 
 | |
2ZU4
 
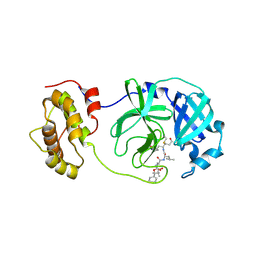 | | Complex structure of SARS-CoV 3CL protease with TG-0204998 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTX
 
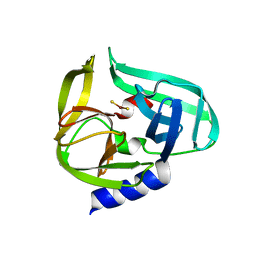 | | Complex structure of CVB3 3C protease with EPDTC | | Descriptor: | 3C proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2Z9J
 
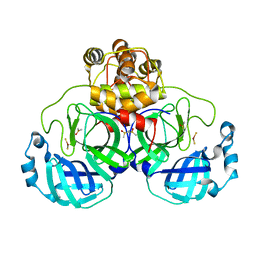 | | Complex structure of SARS-CoV 3C-like protease with EPDTC | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZU5
 
 | | complex structure of SARS-CoV 3CL protease with TG-0205486 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-[(1R)-4-cyclopropyl-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}butyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZU1
 
 | |
1X06
 
 | | Crystal structure of undecaprenyl pyrophosphate synthase in complex with Mg, IPP and Fspp | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
1X07
 
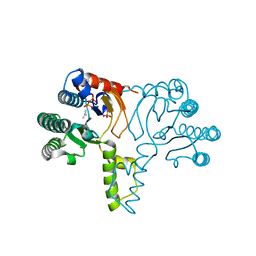 | | Crystal structure of undecaprenyl pyrophosphate synthase in complex with Mg and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
