6NPF
 
 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | Descriptor: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6BFZ
 
 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6BFY
 
 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
7R6G
 
 | | Crystal structure of DfrA5 dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | Dihydrofolate reductase type 5, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7MYM
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7NAE
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6D3Q
 
 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | Deposit date: | 2018-04-16 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
3Q51
 
 | |
3Q50
 
 | |
8VTC
 
 | |
2P7F
 
 | | The Novel Use of a 2',5'-Phosphodiester Linkage as a Reaction Intermediate at the Active Site of a Small Ribozyme | | Descriptor: | COBALT HEXAMMINE(III), Loop A ribozyme strand, Loop B S-turn strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
2P7E
 
 | | Vanadate at the Active Site of a Small Ribozyme Suggests a Role for Water in Transition-State Stabilization | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
2P7D
 
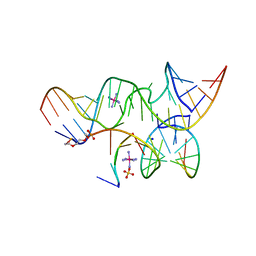 | | A Minimal, 'Hinged' Hairpin Ribozyme Construct Solved with Mimics of the Product Strands at 2.25 Angstroms Resolution | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
3GS8
 
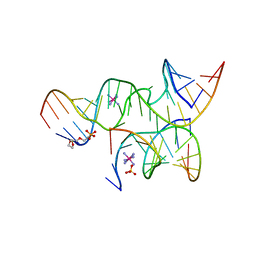 | | An all-RNA hairpin ribozyme A38N1dA38 variant with a transition-state mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GS1
 
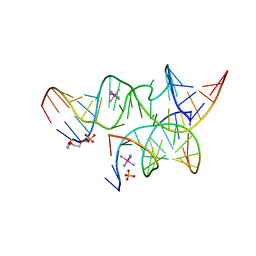 | | An all-RNA Hairpin Ribozyme with mutation A38N1dA | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3CQS
 
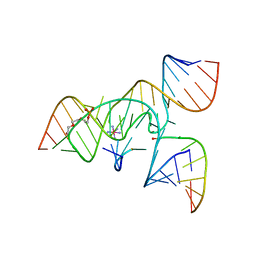 | | A 3'-OH, 2',5'-phosphodiester substitution in the hairpin ribozyme active site reveals similarities with protein ribonucleases | | Descriptor: | 13-mer substrate strand with 3'-OH, 2',5'-phosphodiester covalently linking 5th and 6th nucleotides, 19-mer ribozyme strand, ... | | Authors: | Torelli, A.T, Spitale, R.C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme
Biochem.Biophys.Res.Commun., 371, 2008
|
|
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZOX
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (monoclinic space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZOY
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3GS5
 
 | | An all-RNA hairpin ribozyme A38N1dA variant with a product mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (25-MER), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
4G6R
 
 | | Minimal Hairpin Ribozyme in the Transition State with G8I Variation | | Descriptor: | Loop A Ribozyme strand, Loop A Substrate strand, Loop B Ribozyme Strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.832 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6P
 
 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G6S
 
 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
3I2R
 
 | | Crystal structure of the hairpin ribozyme with a 2',5'-linked substrate with N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2Q
 
 | | Crystal structure of the hairpin ribozyme with 2'OMe substrate strand and N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
