4IGS
 
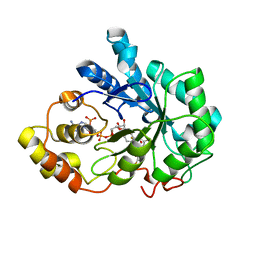 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-18 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IPP
 
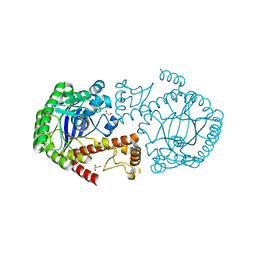 | |
3EGK
 
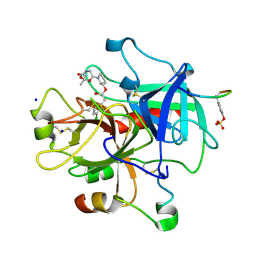 | | KNOBLE Inhibitor | | Descriptor: | Hirudin variant-1, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Baum, B, Heine, A, Klebe, G, Muenzel, M. | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | KNOBLE: a knowledge-based approach for the design and synthesis of readily accessible small-molecule chemical probes to test protein binding
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3EIM
 
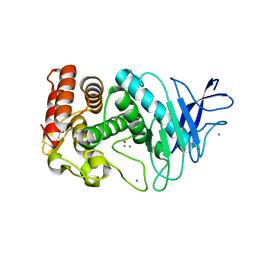 | |
3EQ0
 
 | | Thrombin Inhibitor | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G, Steinmetzer, T. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3F68
 
 | | Thrombin Inhibition | | Descriptor: | Hirudin variant-2, N-acetyl-3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)-L-prolinamide, Prothrombin, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-11-05 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin.
J.Mol.Biol., 391, 2009
|
|
4JBR
 
 | |
3F2P
 
 | | Thermolysin inhibition | | Descriptor: | 3-methyl-2-(propanoyloxy)benzoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
1Y3Y
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YL-1-CYCLOPROPYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, CALCIUM ION, Trypsin, ... | | Authors: | Fokkens, J, Klebe, G. | | Deposit date: | 2004-11-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3F28
 
 | | Thermolysin inhibition | | Descriptor: | 2-[(cyclopropylcarbonyl)oxy]-3-methylbenzoic acid, CALCIUM ION, Thermolysin, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
1Y3U
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YLMETHYL-1-ISOPROPYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, CALCIUM ION, Trypsin, ... | | Authors: | Fokkens, J, Obst, U, Heine, A, Diederich, F, Klebe, G. | | Deposit date: | 2004-11-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Thermodynamic Characterisation of Protein-Inhibitor-Complexes of Thrombin and Trypsin Inhibitors
To be Published
|
|
3FCQ
 
 | | Thermolysin inhibition | | Descriptor: | 2-(acetyloxy)-3-methylbenzoic acid, CALCIUM ION, Thermolysin, ... | | Authors: | Steuber, H, Englert, L, Silber, K, Heine, A, Klebe, G. | | Deposit date: | 2008-11-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
3FB0
 
 | |
3FBO
 
 | |
1Y3W
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YLMETHYL-1-ETHYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, CALCIUM ION, Trypsinogen, ... | | Authors: | Fokkens, J, Klebe, G. | | Deposit date: | 2004-11-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1Y5A
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2-O-{4-[AMINO(IMINO)METHYL]PHENYL}-5-O-{3-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5B
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y3V
 
 | | Trypsin Inhibitor Complex | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YL-1-CYCLOPROPYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, CALCIUM ION, Trypsin, ... | | Authors: | Fokkens, J, Klebe, G. | | Deposit date: | 2004-11-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1Y59
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2,5-BIS-O-{3-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
1Y5U
 
 | | Dianhydrosugar-based benzamidine, factor Xa specific inhibitor in complex with bovine trypsin mutant | | Descriptor: | 2-O-{3-[AMINO(IMINO)METHYL]PHENYL}-5-O-{4-[AMINO(IMINO)METHYL]PHENYL}-1,4:3,6-DIANHYDRO-D-GLUCITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Di Fenza, A, Heine, A, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding binding selectivity toward trypsin and factor Xa: the role of aromatic interactions
Chemmedchem, 2, 2007
|
|
3FLF
 
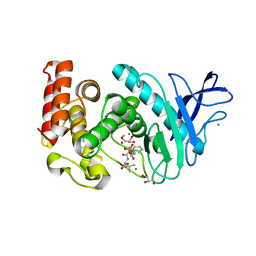 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, GLYCEROL, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-valyl-L-leucine, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2008-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: binding of phosphonamidate to the S1'-pocket of thermolysin.
Biochim.Biophys.Acta, 1800, 2010
|
|
1Y3X
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-{5-(PHENYL[1,3]DIOXOL-5-YLMETHYL)-4-ETHYL-2,3,3-TRIMETHYL-6-OXO-OCTAHYDRO-PYRROLO[3,4-C]PYRROL-1-YL}-BENZAMIDINE, CALCIUM ION, Trypsinogen, ... | | Authors: | Fokkens, J, Klebe, G. | | Deposit date: | 2004-11-26 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1YPE
 
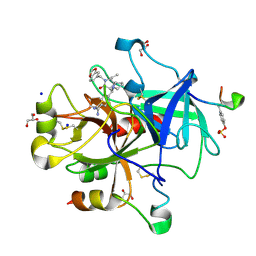 | | Thrombin Inhibitor Complex | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YLMETHYL-1-ETHYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, GLYCEROL, Hirudin, ... | | Authors: | Fokkens, J, Obst-Sander, U, Heine, A, Diederich, F, Klebe, G. | | Deposit date: | 2005-01-31 | | Release date: | 2006-01-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3RM0
 
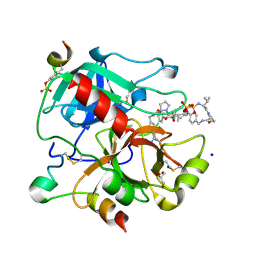 | | Human Thrombin in complex with MI354 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3RMM
 
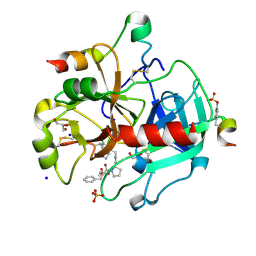 | | Human Thrombin in complex with MI332 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
