4O9W
 
 | |
6A94
 
 | | Crystal structure of 5-HT2AR in complex with zotepine | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(3-chloranylbenzo[b][1]benzothiepin-5-yl)oxy-N,N-dimethyl-ethanamine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6A93
 
 | | Crystal structure of 5-HT2AR in complex with risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5C16
 
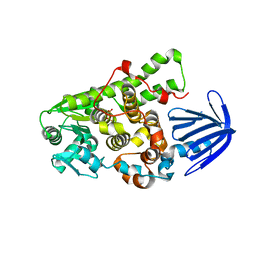 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
3WUU
 
 | |
7WRS
 
 | |
7WRU
 
 | |
4K1V
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y57F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
4K1U
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y32F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LAV
 
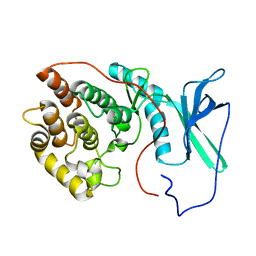 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
8XUY
 
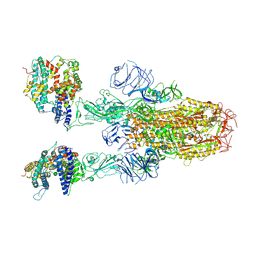 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XVM
 
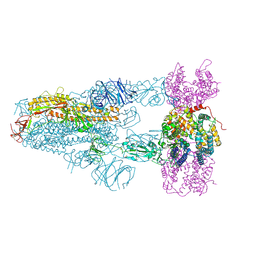 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
9IU1
 
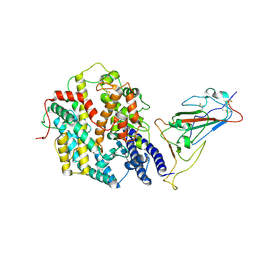 | | Structure of SARS-CoV-2 JN.1 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-07-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUX
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike protein (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV0
 
 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV1
 
 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUZ
 
 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up and 1-down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
7D7M
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
8WXL
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
6JZH
 
 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
