8K6U
 
 | |
8K6X
 
 | | Crystal structure of E.coli Cyanase complex with cyanate and bicarbonate | | 分子名称: | CARBONATE ION, Cyanate hydratase, SULFATE ION, ... | | 著者 | Kim, J, Nam, K.H, Cho, Y. | | 登録日 | 2023-07-25 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8K6G
 
 | |
8K6S
 
 | |
8K6H
 
 | |
8GUY
 
 | | human insulin receptor bound with two insulin molecules | | 分子名称: | Insulin A chain, Insulin, isoform 2, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
4PZD
 
 | |
8JQV
 
 | |
8H26
 
 | |
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | 著者 | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | 登録日 | 2019-10-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
7YQ6
 
 | | human insulin receptor bound with A62 DNA aptamer | | 分子名称: | IR-A62 aptamer, Isoform Short of Insulin receptor | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
7YQ3
 
 | | human insulin receptor bound with A43 DNA aptamer and insulin | | 分子名称: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
7YQ4
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
7YQ5
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin | | 分子名称: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-08-05 | | 公開日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures
Nat Commun, 13, 2022
|
|
5HZ2
 
 | | Crystal structure of PhaC1 from Ralstonia eutropha | | 分子名称: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | 著者 | Kim, J, Kim, K.-J. | | 登録日 | 2016-02-02 | | 公開日 | 2016-12-07 | | 最終更新日 | 2017-04-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
4QNX
 
 | | Crystal structure of apo-CmoB | | 分子名称: | SULFATE ION, tRNA (mo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-06-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.619 Å) | | 主引用文献 | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4QNV
 
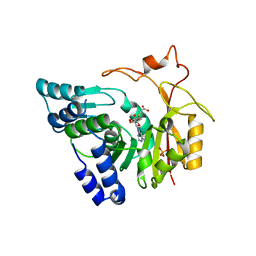 | | Crystal structure of Cx-SAM bound CmoB from E. coli in P6122 | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-06-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4QNU
 
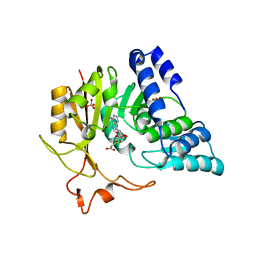 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-06-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
1MIO
 
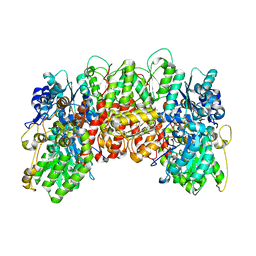 | | X-RAY CRYSTAL STRUCTURE OF THE NITROGENASE MOLYBDENUM-IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM AT 3.0 ANGSTROMS RESOLUTION | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | 著者 | Kim, J, Woo, D, Rees, D.C. | | 登録日 | 1993-03-24 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.
Biochemistry, 32, 1993
|
|
8EBT
 
 | |
8EBS
 
 | |
8EBW
 
 | |
8EBV
 
 | |
8EBX
 
 | |
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | 分子名称: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | 著者 | Kim, J, Yang, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-05-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
