6JQ7
 
 | |
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIG
 
 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
1DU3
 
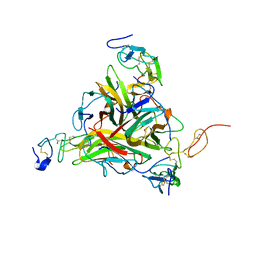 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
8SGH
 
 | |
8TH1
 
 | |
2MJ6
 
 | |
7S5O
 
 | |
5Y7K
 
 | | Crystal structure of human DPP4 in complex with inhibitor1 | | Descriptor: | (R)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazine-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7J
 
 | | Crystal structure of human DPP4 in complex with inhibitor2 | | Descriptor: | (S)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7H
 
 | | Crystal structure of human DPP4 in complex with inhibitor3 | | Descriptor: | (R)-4-(3-amino-4-(2,4,5-trifluorophenyl)butanoyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
1CVM
 
 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
8DLF
 
 | |
1DMQ
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
6DCX
 
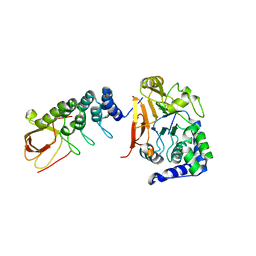 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
7XLJ
 
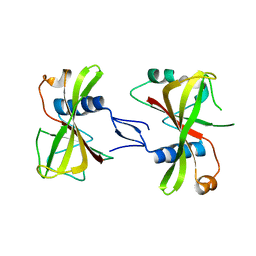 | | The crystal structure of ORE1(ANAC092) NAC domain | | Descriptor: | NAC domain-containing protein 92 | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
7XP3
 
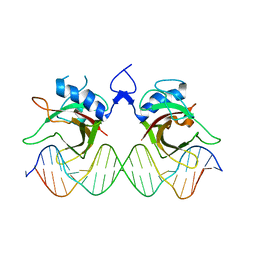 | | DNA complex form of ORESARA1(ANAC092) NAC Domain | | Descriptor: | DNA (5'-D(*AP*GP*TP*TP*AP*CP*GP*TP*AP*CP*GP*GP*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*CP*GP*TP*GP*TP*GP*CP*CP*GP*TP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-05-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
2POO
 
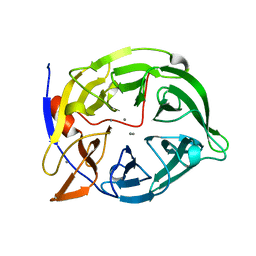 | | THERMOSTABLE PHYTASE IN FULLY CALCIUM LOADED STATE | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
5XV9
 
 | |
6LVP
 
 | |
5Y86
 
 | | Crystal structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | Authors: | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
6LVO
 
 | |
