6C0U
 
 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
3PVB
 
 | | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Boettcher, A.J, Wu, J, Kim, C, Yang, J, Bruystens, J, Cheung, N, Pennypacker, J.K, Blumenthal, D.A, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2010-12-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase
Structure, 19, 2011
|
|
2HWN
 
 | |
4OJK
 
 | | Structure of the cGMP Dependent Protein Kinase II and Rab11b Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11B, cGMP-dependent protein kinase 2 | | Authors: | Reger, A.S, Yang, M.P, Guo, E, Kim, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Crystal Structure of the cGMP-dependent Protein Kinase II Leucine Zipper and Rab11b Protein Complex Reveals Molecular Details of G-kinase-specific Interactions.
J.Biol.Chem., 289, 2014
|
|
2K9J
 
 | | Integrin alphaIIb-beta3 transmembrane complex | | Descriptor: | Integrin alpha-IIb light chain, Integrin beta-3 | | Authors: | Lau, T, Kim, C, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2008-10-15 | | Release date: | 2009-03-24 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling
Embo J., 28, 2009
|
|
2L8E
 
 | |
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
3PNA
 
 | | Crystal Structure of cAMP bound (91-244)RIa Subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Kim, C, Taylor, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Cyclic AMP Analog Blocks Kinase Activation by Stabilizing Inactive Conformation: Conformational Selection Highlights a New Concept in Allosteric Inhibitor Design.
Mol Cell Proteomics, 10, 2011
|
|
2L91
 
 | |
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4Y4V
 
 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4RSI
 
 | | Yeast Smc2-Smc4 hinge domain with extended coiled coils | | Descriptor: | PHOSPHATE ION, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Soh, Y.M, Shin, H.C, Oh, B.H. | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for SMC Rod Formation and Its Dissolution upon DNA Binding.
Mol.Cell, 57, 2015
|
|
4RSJ
 
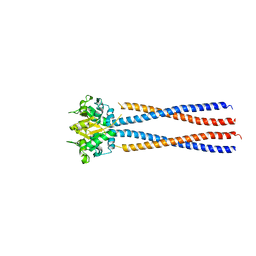 | |
5DYL
 
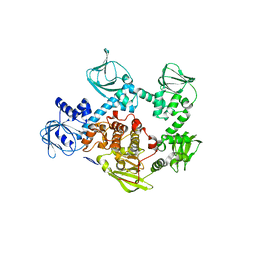 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - Apo form | | Descriptor: | cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DYK
 
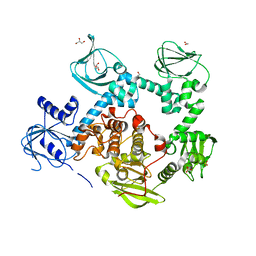 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum - Apo form | | Descriptor: | 1,2-ETHANEDIOL, CGMP-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5X1E
 
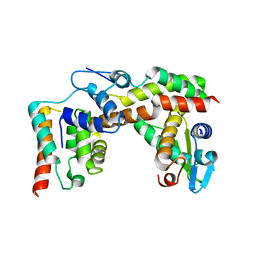 | |
5X1U
 
 | |
5E16
 
 | | Co-crystal structure of the N-termial cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DZC
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - AMPPNP bound | | Descriptor: | CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, He, H, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5X1H
 
 | | Structure of Legionella pneumophila DotN | | Descriptor: | IcmJ (DotN), ZINC ION | | Authors: | Kwak, M.J, Oh, B.H. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
7MBJ
 
 | |
7BYP
 
 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7BYO
 
 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D01
 
 | | Lysozyme structure SS2 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7D04
 
 | | Lysozyme structure SS3 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
