1SI7
 
 | | Structure of E. coli tRNA psi 13 pseudouridine synthase TruD | | Descriptor: | tRNA pseudouridine synthase D | | Authors: | Kaya, Y, Del Campo, M, Ofengand, J, Malhotra, A. | | Deposit date: | 2004-02-27 | | Release date: | 2004-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TruD, a novel pseudouridine synthase with a new protein fold
J.Biol.Chem., 279, 2004
|
|
3AW6
 
 | |
3AW7
 
 | |
1I2A
 
 | |
3IVK
 
 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
1K9A
 
 | | Crystal structure analysis of full-length carboxyl-terminal Src kinase at 2.5 A resolution | | Descriptor: | Carboxyl-terminal Src kinase | | Authors: | Ogawa, A, Takayama, Y, Nagata, A, Chong, K.T, Takeuchi, S, Sakai, H, Nakagawa, A, Nada, S, Okada, M, Tsukihara, T. | | Deposit date: | 2001-10-28 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the carboxyl-terminal Src kinase, Csk.
J.Biol.Chem., 277, 2002
|
|
2DGE
 
 | | Crystal structure of oxidized cytochrome C6A from Arabidopsis thaliana | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Chida, H, Yokoyama, T, Kawai, F, Nakazawa, A, Akazaki, H, Takayama, Y, Hirano, T, Suruga, K, Satoh, T, Yamada, S, Kawachi, R, Unzai, S, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2006-03-11 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of oxidized cytochrome c(6A) from Arabidopsis thaliana
Febs Lett., 580, 2006
|
|
2CVC
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
8Z56
 
 | |
8Z55
 
 | |
8Z58
 
 | |
8Z54
 
 | |
8Z57
 
 | |
6U8D
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | Descriptor: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
2ROA
 
 | | Solution structure of calcium bound soybean calmodulin isoform 4 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO9
 
 | | Solution structure of calcium bound soybean calmodulin isoform 1 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin-2 | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2ROB
 
 | | Solution structure of calcium bound soybean calmodulin isoform 4 C-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
2RO8
 
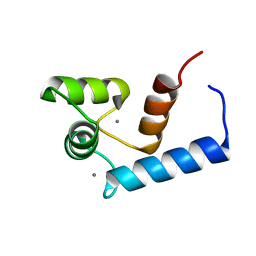 | | Solution structure of calcium bound soybean calmodulin isoform 1 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
4KZD
 
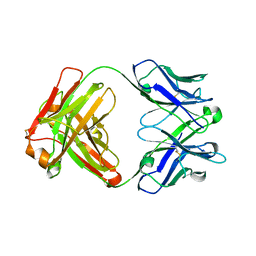 | | Crystal structure of an RNA aptamer in complex with fluorophore and Fab | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, BL3-6 Fab antibody, heavy chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4KZE
 
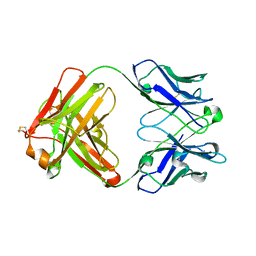 | | Crystal structure of an RNA aptamer in complex with Fab | | Descriptor: | BL3-6 Fab antibody, heavy chain, light chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
2KX9
 
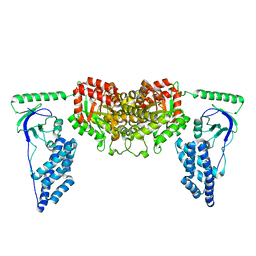 | | Solution Structure of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Schwieters, C.D, Suh, J, Grishaev, A, Takayama, Y, Guirlando, R, Clore, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2LOE
 
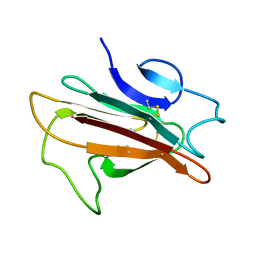 | | Structure of the Plasmodium 6-cysteine s48/45 Domain | | Descriptor: | 6-cysteine protein, putative | | Authors: | Cai, M, Arredondo, S.A, Clore, M.G, Miller, L.H, Takayama, Y, Macdonald, N.J, Enderson, E.D, Aravind, L. | | Deposit date: | 2012-01-23 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Plasmodium 6-cysteine s48/45 domain.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4Q9Q
 
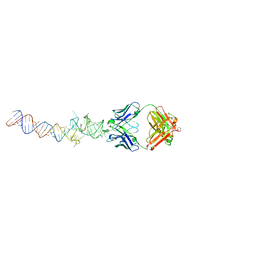 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
2XDF
 
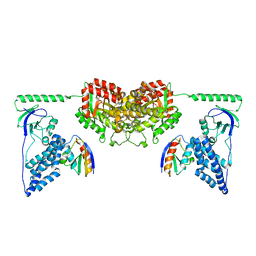 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
