6JND
 
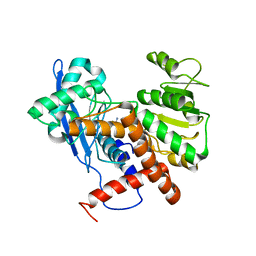 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6KFK
 
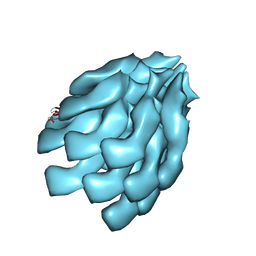 | |
6LKT
 
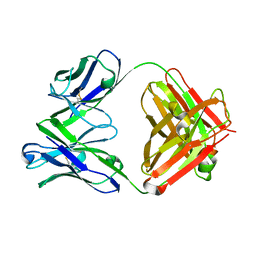 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | Descriptor: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6LTG
 
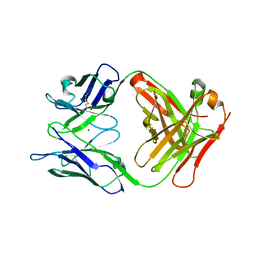 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
1WUJ
 
 | | Three-Dimensional Structure Of The Ni-B State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUH
 
 | | Three-Dimensional Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis for the activation process of [NiFe] hydrogenase from D.vulgaris Miyazaki F
To be Published
|
|
1WUK
 
 | | High resolution Structure Of The Oxidized State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUL
 
 | | High Resolution Structure Of The Reduced State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUI
 
 | | Ultra-High resolution Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
6LY9
 
 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6LY8
 
 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
3VXV
 
 | | Crystal structure of methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*CP*(5CM)P*GP*GP*AP*CP*A)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VXX
 
 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/5mCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5CM)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYQ
 
 | | Crystal structure of the methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence in space group P1 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*AP*TP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYB
 
 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/hmCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5HC)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
7W71
 
 | |
7W6Y
 
 | | Crystal structure of Kangiella koreensis RseP orthologue in complex with batimastat in space group P1 | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, Anti sigma-E protein, RseA, ... | | Authors: | Imaizumi, Y, Takanuki, K, Nogi, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP.
Sci Adv, 8, 2022
|
|
7W6X
 
 | |
7W6Z
 
 | |
7W70
 
 | |
1IS1
 
 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
8H4M
 
 | | Crystal Structure of GTP-bound Irgb6_T95D mutant | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
8H4O
 
 | | Crystal Structure of nucleotide-free Irgb6_T95D mutant | | Descriptor: | T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
6E0R
 
 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-07-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
