5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5AZ2
 
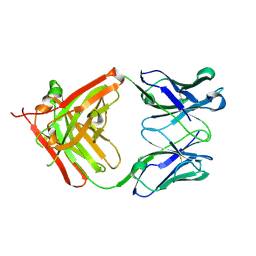 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
3VI7
 
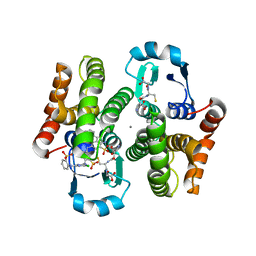 | |
3VI5
 
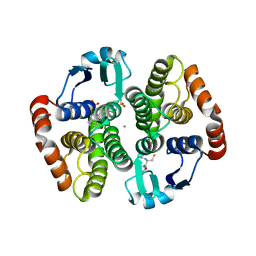 | | Human hematopoietic prostaglandin D synthase inhibitor complex structures | | Descriptor: | 1-amino-9,10-dioxo-4-[(4-sulfamoylphenyl)amino]-9,10-dihydroanthracene-2-sulfonic acid, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kado, Y, Inoue, T. | | Deposit date: | 2011-09-21 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human hematopoietic prostaglandin D synthase inhibitor complex structures
J.Biochem., 151, 2012
|
|
3APG
 
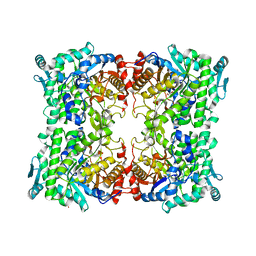 | |
1V40
 
 | | First Inhibitor Complex Structure of Human Hematopoietic Prostaglandin D Synthase | | Descriptor: | 3-(1,3-BENZOTHIAZOL-2-YL)-2-(1,4-DIOXO-1,2,3,4-TETRAHYDROPHTHALAZIN-6-YL)-5-[(E)-2-PHENYLVINYL]-3H-TETRAAZOL-2-IUM, GLUTATHIONE, GLYCEROL, ... | | Authors: | Inoue, T, Okano, Y, Kado, Y, Aritake, K, Irikura, D, Uodome, N, Kinugasa, S, Okazaki, N, Matsumura, H, Kai, Y, Urade, Y. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First determination of the inhibitor complex structure of human hematopoietic prostaglandin D synthase.
J.Biochem.(Tokyo), 135, 2004
|
|
2CVD
 
 | | Crystal structure analysis of human hematopoietic prostaglandin D synthase complexed with HQL-79 | | Descriptor: | 4-(BENZHYDRYLOXY)-1-[3-(1H-TETRAAZOL-5-YL)PROPYL]PIPERIDINE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Aritake, K, Kado, Y, Inoue, T, Miyano, M, Urade, Y. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of HQL-79, an Orally Selective Inhibitor of Human Hematopoietic Prostaglandin D Synthase.
J.Biol.Chem., 281, 2006
|
|
6A77
 
 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the Fab fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, Roundabout homolog 1 | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A76
 
 | | Crystal structure of the Fab fragment of B5209B, a murine monoclonal antibody specific for the fifth immunoglobulin domain (Ig5) of human ROBO1 | | Descriptor: | GLYCEROL, Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A78
 
 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the scFv fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain and linker region of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A79
 
 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the mutant scFv fragment (P103A) of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Yokota, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
3WIH
 
 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WII
 
 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | Descriptor: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WO8
 
 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | Beta-N-acetylglucosaminidase | | Authors: | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
3A2W
 
 | | Peroxiredoxin (C50S) from Aeropytum pernix K1 (peroxide-bound form) | | Descriptor: | GLYCEROL, PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A2V
 
 | | Peroxiredoxin (C207S) from Aeropyrum pernix K1 complexed with hydrogen peroxide | | Descriptor: | PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Ishikawa, K, Matsumura, H, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A2X
 
 | | Peroxiredoxin (C50S) from Aeropyrum pernix K1 (acetate-bound form) | | Descriptor: | ACETATE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
5NEN
 
 | |
5X7K
 
 | |
3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
3AFB
 
 | |
8I2Q
 
 | | Beijerinckia indica beta-fructosyltransferase variant H395R/F473Y | | Descriptor: | Beta-fructosyltransferase, GLYCEROL | | Authors: | Tonozuka, T. | | Deposit date: | 2023-01-15 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization and alteration of product specificity of Beijerinckia indica subsp. indica beta-fructosyltransferase.
Biosci.Biotechnol.Biochem., 87, 2023
|
|
8I2R
 
 | |
