1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
8V2T
 
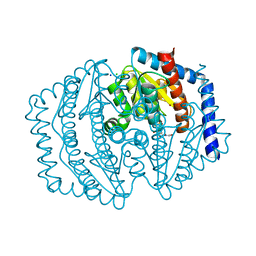 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148591 | | Descriptor: | 1,5,6-trideoxy-6,6-difluoro-1-(N-hydroxyformamido)-6-phosphono-D-ribo-hexitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
8V4J
 
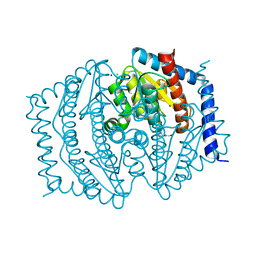 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
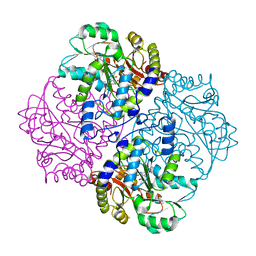
2FQ6
 
 | |
1EA6
 
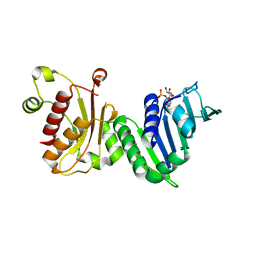 | |
6ED7
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772 | | Descriptor: | 2-[(2-nitrophenyl)sulfanyl]acetohydrazide, 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Brown, C.M, Zlitni, S, Chan, J, Brown, E.D, Junop, M.S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772
To Be Published
|
|
2OPT
 
 | | Crystal Structure of Apo ActR from Streptomyces coelicolor. | | Descriptor: | ActII protein | | Authors: | Willems, A.R, Junop, M.S. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
8U0G
 
 | |
8U1J
 
 | |
2R9A
 
 | | Crystal structure of human XLF | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Mol.Cell, 28
|
|
7UDI
 
 | |
3B6A
 
 | | Crystal structure of the Streptomyces coelicolor TetR family protein ActR in complex with actinorhodin | | Descriptor: | 2,2'-[(1R,1'R,3S,3'S)-6,6',9,9'-tetrahydroxy-1,1'-dimethyl-5,5',10,10'-tetraoxo-3,3',4,4',5,5',10,10'-octahydro-1H,1'H-8,8'-bibenzo[g]isochromene-3,3'-diyl]diacetic acid, ActR protein | | Authors: | Willems, A.R, Junop, M.S. | | Deposit date: | 2007-10-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
3B6C
 
 | |
4NOA
 
 | |
4NOE
 
 | | Crystal structure of DdrB bound to 30b ssDNA | | Descriptor: | 5'-D(*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP)-3', CALCIUM ION, Single-stranded DNA-binding protein DdrB | | Authors: | Sugiman-Marangos, S.N, Weiss, Y.M, Junop, M.S. | | Deposit date: | 2013-11-19 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4NPH
 
 | | Crystal structure of SsaN from Salmonella enterica | | Descriptor: | Probable secretion system apparatus ATP synthase SsaN | | Authors: | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
6PPW
 
 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with magnesium and malate | | Descriptor: | D-MALATE, MAGNESIUM ION, N-acetylneuraminate synthase | | Authors: | Rosanally, A.Z, Junop, M.S, Berti, P.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
6PPX
 
 | |
6PPZ
 
 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with manganese, inorganic phosphate, and N-acetylmannosamine (NeuB.Mn2+.Pi.ManNAc) | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, MANGANESE (II) ION, N-acetylneuraminate synthase, ... | | Authors: | Berti, P.J, Junop, M.S. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
3Q9D
 
 | |
3RWR
 
 | | Crystal structure of the human XRCC4-XLF complex | | Descriptor: | DNA repair protein XRCC4, HEXATANTALUM DODECABROMIDE, Non-homologous end-joining factor 1 | | Authors: | Andres, S.N, Junop, M.S. | | Deposit date: | 2011-05-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.943 Å) | | Cite: | Structure of human XLF-XRCC4: assembly of a functional DNA repair complex
To be Published
|
|
1H7S
 
 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1H7U
 
 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
2GMW
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli. | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Zhang, K, DeLeon, G, Wright, G.D, Junop, M.S. | | Deposit date: | 2006-04-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
2GQN
 
 | |
