9E3A
 
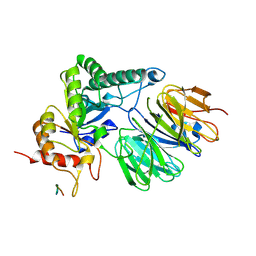 | |
9E3C
 
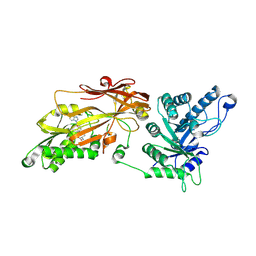 | |
9E3B
 
 | | Cryo-EM structure of PRMT5/WDR77 in complex with 6S complex | | Descriptor: | Methylosome protein WDR77, Methylosome subunit pICln, Protein arginine N-methyltransferase 5, ... | | Authors: | Jin, C.Y, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2024-10-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Substrate adaptors are flexible tethering modules that enhance substrate methylation by the arginine methyltransferase PRMT5.
J.Biol.Chem., 301, 2025
|
|
8G46
 
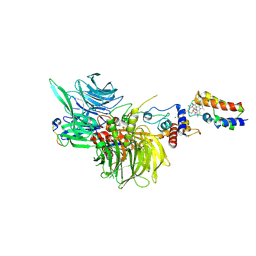 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
7MWD
 
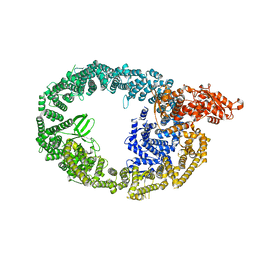 | | HUWE1 in map with focus on HECT | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MWF
 
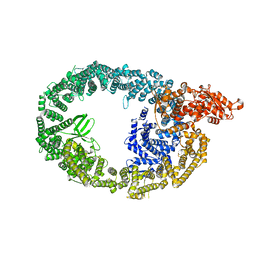 | | HUWE1 in map with focus on interface | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MOP
 
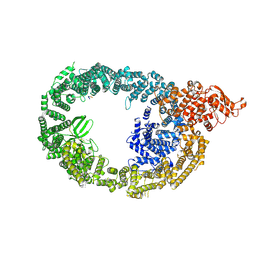 | | Cryo-EM structure of human HUWE1 in complex with DDIT4 | | Descriptor: | DNA damage-inducible transcript 4 protein, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MWE
 
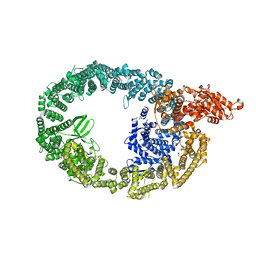 | | HUWE1 in map with focus on WWE | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
8E2J
 
 | | Cryo-EM structure of BIRC6/Smac (from local refinement 1) | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Diablo IAP-binding mitochondrial protein | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2D
 
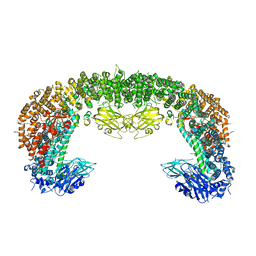 | | Cryo-EM structure of BIRC6 (consensus) | | Descriptor: | Baculoviral IAP repeat-containing protein 6 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2G
 
 | |
8E2E
 
 | |
8E2I
 
 | | Cryo-EM structure of BIRC6/Smac | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Diablo IAP-binding mitochondrial protein | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2F
 
 | |
8E2K
 
 | | Cryo-EM structure of BIRC6/HtrA2-S306A | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases.
Science, 379, 2023
|
|
8E2H
 
 | |
7JQ9
 
 | | Cryo-EM structure of human HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2020-08-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
