1SJT
 
 | | MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1SJU
 
 | | MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1VKT
 
 | | HUMAN INSULIN TWO DISULFIDE MODEL, NMR, 10 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Frank, B.H, Jia, W.H, Chu, Y.C, Wang, S.H, Burke, G.T, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1996-10-14 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue.
J.Mol.Biol., 264, 1996
|
|
6NO9
 
 | | PIM1 in complex with Cpd16 (5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide) | | Descriptor: | 5-amino-N-{5-[(4R,5R)-4-amino-5-fluoroazepan-1-yl]-1-methyl-1H-pyrazol-4-yl}-2-(2,6-difluorophenyl)-1,3-thiazole-4-carboxamide, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Optimization of Pan-Pim Kinase Activity and Oral Bioavailability Leading to Diaminopyrazole (GDC-0339) for the Treatment of Multiple Myeloma.
J. Med. Chem., 62, 2019
|
|
8WO7
 
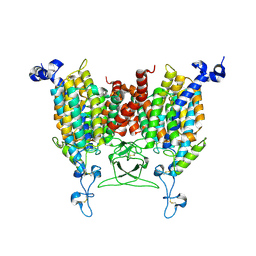 | | Apo state of Arabidopsis AZG1 T440Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8WMQ
 
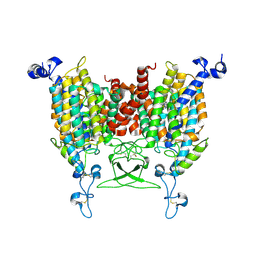 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH5.5 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
3BXQ
 
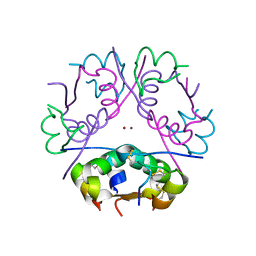 | | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition | | Descriptor: | ZINC ION, insulin A chain, insulin B chain | | Authors: | Wan, Z.L, Huang, K, Hu, S.Q, Whittaker, J, Weiss, M.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition.
J.Biol.Chem., 283, 2008
|
|
3D34
 
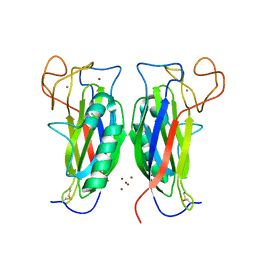 | | Structure of the F-spondin domain of mindin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Spondin-2 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-02-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Embo J., 28, 2009
|
|
3DCV
 
 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | Descriptor: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
8E4W
 
 | |
8E5C
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L50F with Nirmatrelvir captured in two conformational states | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8DT9
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
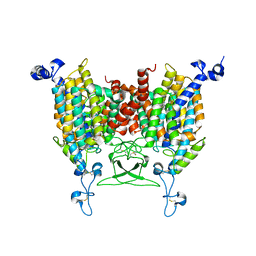 | | 6-BAP bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
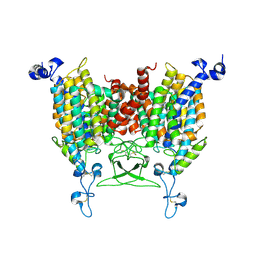 | | kinetin bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
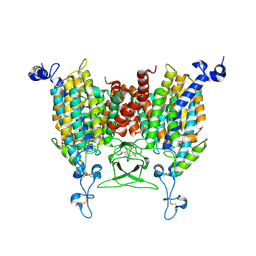 | | Endogenous substrate adenine bound state of Arabidopsis AZG1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRO
 
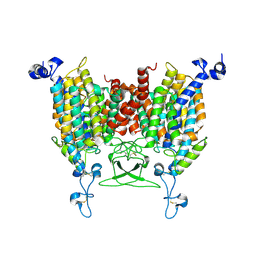 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
5ZXV
 
 | |
3FQ9
 
 | | Design of an insulin analog with enhanced receptor-binding selectivity. Rationale, structure, and therapeutic implications | | Descriptor: | Insulin, ZINC ION | | Authors: | Zhao, M, Wan, Z.L, Whittaker, L, Xu, B, Phillips, N, Katsoyannis, P, Whittaker, J, Weiss, M.A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of an insulin analog with enhanced receptor binding selectivity: rationale, structure, and therapeutic implications.
J.Biol.Chem., 284, 2009
|
|
3GKY
 
 | | The Structural Basis of an ER Stress-Associated Bottleneck in a Protein Folding Landscape | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Liu, M, Wan, Z.L, Chu, Y.C, Alddin, H, Klaproth, B, Weiss, M.A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a "nonfoldable" insulin: IMPAIRED FOLDING EFFICIENCY DESPITE NATIVE ACTIVITY.
J.Biol.Chem., 284, 2009
|
|
1JB6
 
 | |
7X2A
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
