8RGG
 
 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
8RGI
 
 | | Structure of DYNLT1:DYNLT2B (TCTEX1:TCTEX1D2) heterodimer. | | Descriptor: | Dynein light chain Tctex-type 1, Dynein light chain Tctex-type protein 2B | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
5NP8
 
 | | PGK1 in complex with CRT0063465 (3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid) | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, 3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid, ... | | Authors: | Turnbull, A.P, Bilsland, A.E, Liu, Y, Sumpton, D, Stevenson, K, Cairney, C.J, Roffey, J, Jenkinson, D, Keith, W.N. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Pyrazolopyrimidine Ligand of Human PGK1 and Stress Sensor DJ1 Modulates the Shelterin Complex and Telomere Length Regulation.
Neoplasia, 21, 2019
|
|
5CZX
 
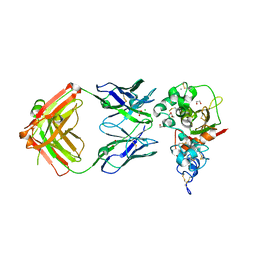 | | Crystal structure of Notch3 NRR in complex with 20358 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 20358 Fab heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
5CZV
 
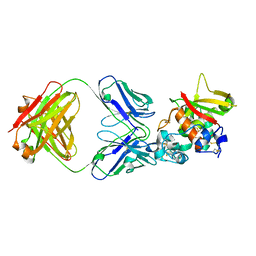 | | Crystal structure of Notch3 NRR in complex with 20350 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 20350 heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
5N4A
 
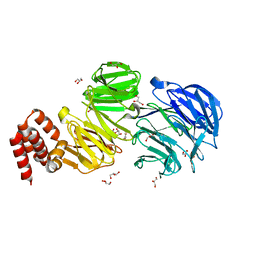 | | Crystal structure of Chlamydomonas IFT80 | | Descriptor: | GLYCEROL, Intraflagellar transport protein 80, OXALATE ION | | Authors: | Taschner, M, Mourao, A. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of intraflagellar transport protein 80 reveals a homo-dimer required for ciliogenesis.
Elife, 7, 2018
|
|
5EW9
 
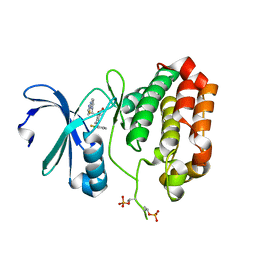 | | Crystal Structure of Aurora A Kinase Domain Bound to MK-5108 | | Descriptor: | 4-(3-chloranyl-2-fluoranyl-phenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid, Aurora kinase A | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | A Cell Biologist's Field Guide to Aurora Kinase Inhibitors.
Front Oncol, 5, 2015
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
