6E9H
 
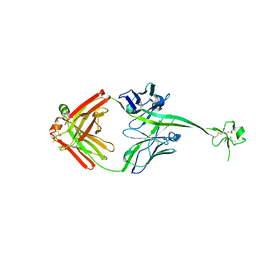 | |
6E8V
 
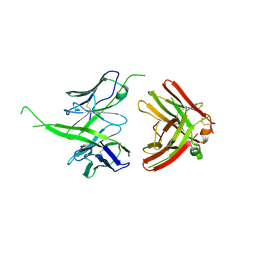 | | The crystal structure of bovine ultralong antibody BOV-1 | | Descriptor: | Bovine ultralong antibody BOV-1 Heavy chain, Bovine ultralong antibody BOV-1 light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2018-07-31 | | Release date: | 2019-09-04 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Front Immunol, 10, 2019
|
|
6E9U
 
 | | The crystal structure of bovine ultralong antibody BOV-7 | | Descriptor: | Bovine ultralong antibody BOV-7 heavy chain, Bovine ultralong antibody BOV-7 light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2018-08-01 | | Release date: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Front Immunol, 10, 2019
|
|
1X9K
 
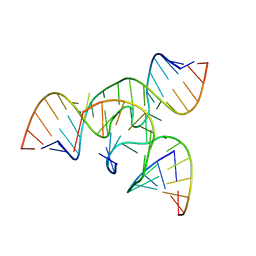 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*AP*AP*UP*AP*GP*AP*GP*AP*AP*GP*CP*GP*A)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*GP*CP*AP*GP*UP*CP*CP*UP*AP*UP*U)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-21 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
6E9K
 
 | |
1H2B
 
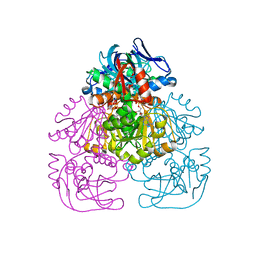 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
6E9Q
 
 | |
1Y6Q
 
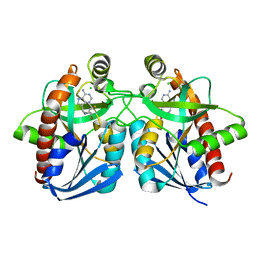 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1Y6R
 
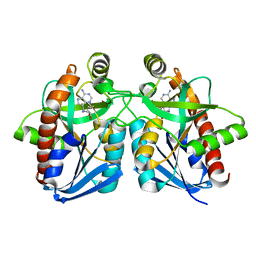 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1Y7Y
 
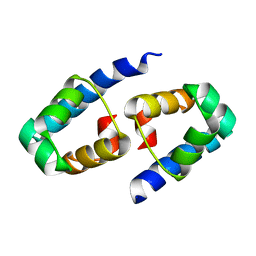 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
1HJK
 
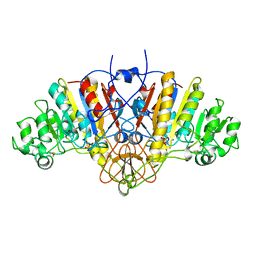 | | ALKALINE PHOSPHATASE MUTANT H331Q | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Murphy, J.E, Stec, B, Ma, L, Kantrowitz, E.R. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trapping and visualization of a covalent enzyme-phosphate intermediate.
Nat.Struct.Biol., 4, 1997
|
|
1YPT
 
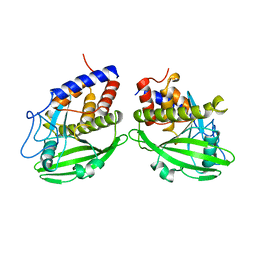 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
1HJG
 
 | | Alteration of the co-substrate selectivity of deacetoxycephalosporin C synthase: The role of arginine-258 | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION | | Authors: | Lee, H.J, Lloyd, M.D, Clifton, I.J, Harlos, K, Dubus, A, Baldwin, J.E, Frere, J.M, Schofield, C.J. | | Deposit date: | 2001-01-15 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of the 2-Oxoacid Cosubstrate Selectivity in Deacetoxycephalosporin C Synthase: The Role of Arginine-258
J.Biol.Chem., 276, 2001
|
|
1HJF
 
 | | Alteration of the co-substrate selectivity of deacetoxycephalosporin C synthase: The role of arginine-258 | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION | | Authors: | Lee, H.J, Lloyd, M.D, Clifton, I.J, Harlos, K, Dubus, A, Baldwin, J.E, Frere, J.M, Schofield, C.J. | | Deposit date: | 2001-01-15 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Alteration of the 2-Oxoacid Cosubstrate Selectivity in Deacetoxycephalosporin C Synthase: The Role of Arginine-258
J.Biol.Chem., 276, 2001
|
|
1J2Y
 
 | | Crystal structure of the type II 3-dehydroquinase | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Lee, B.I, Kwak, J.E, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the type II 3-dehydroquinase from Helicobacter pylori
Proteins, 51, 2003
|
|
1ZA7
 
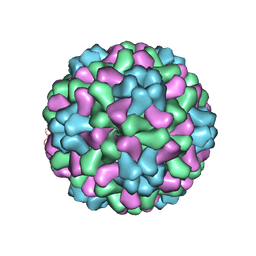 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
1YVL
 
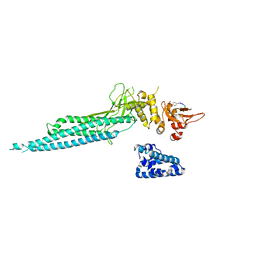 | | Structure of Unphosphorylated STAT1 | | Descriptor: | 5-residue peptide, GOLD ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Mao, X, Ren, Z, Parker, G.N, Sondermann, H, Pastorello, M.A, Wang, W, McMurray, J.S, Demeler, B, Darnell Jr, J.E, Chen, X. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of unphosphorylated STAT1 association and receptor binding.
Mol.Cell, 17, 2005
|
|
1IF0
 
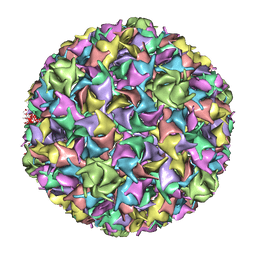 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1IPS
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (MANGANESE COMPLEX) | | Descriptor: | ISOPENICILLIN N SYNTHASE, MANGANESE (II) ION | | Authors: | Roach, P.L, Clifton, I.J, Fulop, V, Harlos, K, Barton, G.J, Hajdu, J, Andersson, I, Schofield, C.J, Baldwin, J.E. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes.
Nature, 375, 1995
|
|
4A5P
 
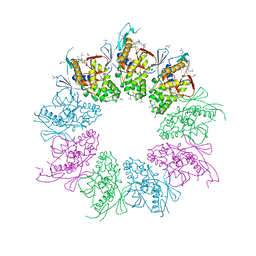 | | Structure of the Shigella flexneri MxiA protein | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN MXIA | | Authors: | Abrusci, P, Vegara-Irigaray, M, Johnson, S, Roversi, P, Friede, M.E, Deane, J.E, Tang, C.M, Lea, S.M. | | Deposit date: | 2011-10-26 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Architecture of the major component of the type III secretion system export apparatus.
Nat.Struct.Mol.Biol., 20, 2013
|
|
203D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
204D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
2ATC
 
 | | CRYSTAL AND MOLECULAR STRUCTURES OF NATIVE AND CTP-LIGANDED ASPARTATE CARBAMOYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN, REGULATORY CHAIN, ... | | Authors: | Honzatko, R.B, Crawford, J.L, Monaco, H.L, Ladner, J.E, Edwards, B.F.P, Evans, D.R, Warren, S.G, Wiley, D.C, Ladner, R.C, Lipscomb, W.N. | | Deposit date: | 1982-03-24 | | Release date: | 1982-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli.
J.Mol.Biol., 160, 1982
|
|
5KEM
 
 | | EBOV sGP in complex with variable Fab domains of IgGs c13C6 and BDBV91 | | Descriptor: | BDBV91 variable Fab domain heavy chain, BDBV91 variable Fab domain light chain, Ebola secreted glycoprotein, ... | | Authors: | Pallesen, J, Murin, C.D, de Val, N, Cottrell, C.A, Hastie, K.M, Turner, H.L, Fusco, M.L, Flyak, A.I, Zeitlin, L, Crowe Jr, J.E, Andersen, K.G, Saphire, E.O, Ward, A.B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of Ebola virus GP and sGP in complex with therapeutic antibodies.
Nat Microbiol, 1, 2016
|
|
7L7E
 
 | |
