4X4G
 
 | |
4X4F
 
 | |
4X4H
 
 | |
4X4D
 
 | |
4X4B
 
 | |
4X4C
 
 | |
5Y8R
 
 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
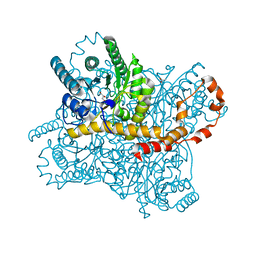 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Z6V
 
 | |
4FN3
 
 | |
4FBI
 
 | |
6UIG
 
 | |
6V3Z
 
 | |
6V4R
 
 | |
6V4Q
 
 | | Crystal structure of a MR78-like antibody naive-1 Fab | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Naive-1 Fab heavy chain, ... | | Authors: | Bozhanova, N.G, Crowe, J.E, Meiler, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of Marburg virus neutralizing antibodies from virus-naive human antibody repertoires using large-scale structural predictions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6APX
 
 | | Crystal structure of human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the monobody YSX1 | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Dual specificity protein phosphatase 1, Monobody YSX1, ... | | Authors: | Gumpena, R, Lountos, G.T, Sreejith, R.K, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Protein Sci., 27, 2018
|
|
6ARK
 
 | | Crystal Structure of compound 10 covalently bound to K-Ras G12C | | Descriptor: | (3R)-N-(6-bromonaphthalen-2-yl)-3-hydroxy-1-propanoyl-L-prolinamide, GLYCEROL, GTPase KRas, ... | | Authors: | Nnadi, C.I, Jenkins, M.L, Gentile, D.R, Bateman, L.A, Zaidman, D, Balius, T.E, Nomura, D.K, Burke, J.E, Shokat, K.M, London, N. | | Deposit date: | 2017-08-22 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange.
J Chem Inf Model, 58, 2018
|
|
6BB4
 
 | |
6B3X
 
 | | Crystal structure of CstF-50 in complex with CstF-77 | | Descriptor: | Cleavage stimulation factor subunit 1, Cleavage stimulation factor subunit 3 | | Authors: | Yang, W, Hsu, P, Yang, F, Song, J.E, Varani, G. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Nucleic Acids Res., 46, 2018
|
|
6BN8
 
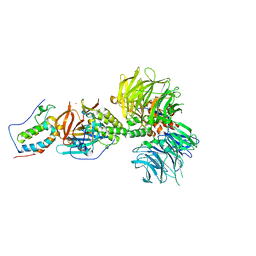 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN9
 
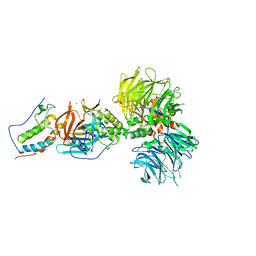 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN7
 
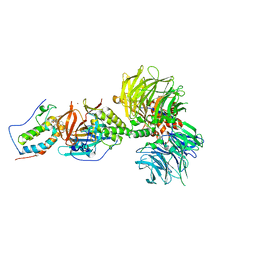 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
