8A0K
 
 | | crystal structure of the kinetoplastid kinetochore protein Trypanosoma brucei KKT3 Divergent Polo-Box domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein kinase, putative | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O.O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
8A0J
 
 | | Crystal structure of the kinetoplastid kinetochore protein Trypanosoma congolense KKT2 divergent polo-box domain | | Descriptor: | Uncharacterized protein TCIL3000_11_11110 | | Authors: | Ishii, M, Ludzia, P, Marciano, G, Allen, W, Nerusheva, O, Akiyoshi, B. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent polo boxes in KKT2 bind KKT1 to initiate the kinetochore assembly cascade in Trypanosoma brucei.
Mol.Biol.Cell, 33, 2022
|
|
6TLX
 
 | | Crystal structure of the unconventional kinetochore protein Perkinsela sp. KKT2a central domain | | Descriptor: | Protein kinase, ZINC ION | | Authors: | Marciano, G, Nerusheva, O, Ishii, M, Akiyoshi, B. | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT2 and KKT3 have unique centromere localization domains.
J.Cell Biol., 220, 2021
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | Descriptor: | kinetochore protein KKT14 | | Authors: | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
6TLY
 
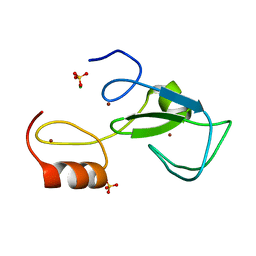 | | Crystal structure of the unconventional kinetochore protein Bodo saltans KKT2 central domain | | Descriptor: | CHLORIDE ION, Protein kinase, putative, ... | | Authors: | Marciano, G, Nerusheva, O, Ishii, M, Akiyoshi, B. | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT2 and KKT3 have unique centromere localization domains.
J.Cell Biol., 220, 2021
|
|
9EVQ
 
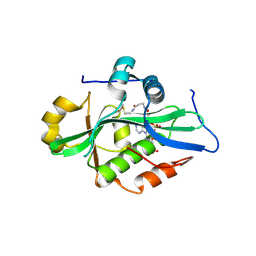 | |
9EVR
 
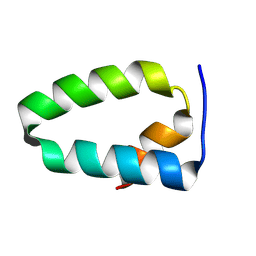 | |
9F5Q
 
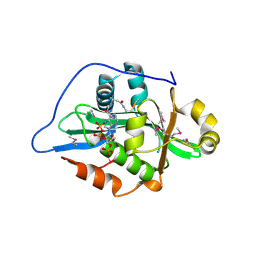 | |
5H6T
 
 | | Crystal structure of Hydrazidase from Microbacterium sp. strain HM58-2 | | Descriptor: | Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
4IJ5
 
 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase from Hydrogenobacter thermophilus TK-6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoserine phosphatase 1 | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
4IJ6
 
 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
5XFA
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
5XF9
 
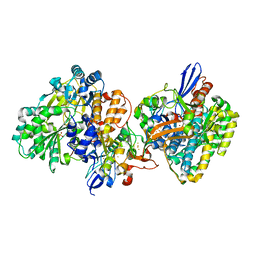 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
8WUC
 
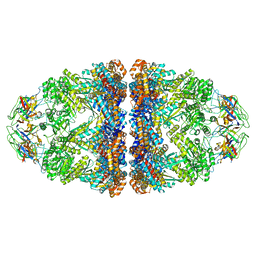 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUX
 
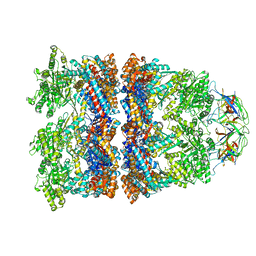 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUW
 
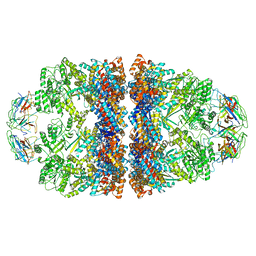 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WU4
 
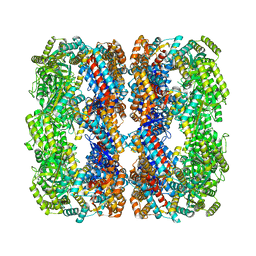 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
5H6S
 
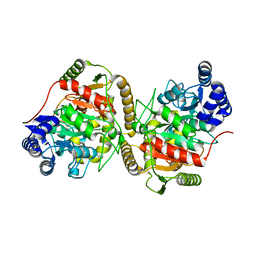 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | Descriptor: | 4-oxidanylbenzohydrazide, Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
6LDY
 
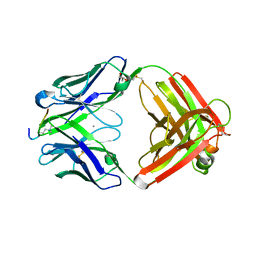 | |
6LDX
 
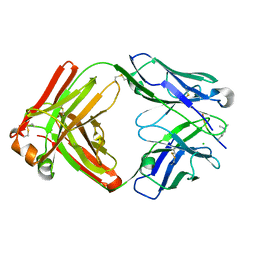 | |
6LDW
 
 | |
6LDV
 
 | |
7DC8
 
 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
1NRR
 
 | |
