4LMH
 
 | | Crystal structure of the outer membrane decaheme cytochrome OmcA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Extracellular iron oxide respiratory system surface decaheme cytochrome c component OmcA, ... | | Authors: | Edwards, M.J, Baiden, N, Clarke, T.A. | | Deposit date: | 2013-07-10 | | Release date: | 2014-04-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into electron transfer at the microbe-mineral interface: the X-ray crystal structures of Shewanella oneidensis MtrC and OmcA
FEBS Letters, 2014
|
|
2B2C
 
 | | Cloning, expression, characterisation and three- dimensional structure determination of the Caenorhabditis elegans spermidine synthase | | Descriptor: | spermidine synthase | | Authors: | Dufe, V.T, Luersen, K, Eschbach, M.L, Haider, N, Karlberg, T, Walter, R.D, Al-Karadaghi, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cloning, expression, characterisation and three-dimensional structure determination of Caenorhabditis elegans spermidine synthase
FEBS LETT., 579, 2005
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
3W1F
 
 | | Crystal structure of Human MPS1 catalytic domain in complex with 5-(5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl)-2-methylbenzenesulfonamide | | Descriptor: | 5-[5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl]-2-methylbenzenesulfonamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Tachibana, Y, Itoh, T, Yamamoto, T, Hashizume, H, Hato, Y, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Higaki, M, Ueda, K, Yoshizawa, H, Baba, Y, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Indazole-based potent and cell-active Mps1 kinase inhibitors: rational design from pan-kinase inhibitor anthrapyrazolone (SP600125)
J.Med.Chem., 56, 2013
|
|
3WZJ
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-(6-(cyclohexylamino)-8-(((tetrahydro-2H-pyran-4-yl)methyl)amino)imidazo[1,2-b]pyridazin-3-yl)-N-cyclopropylbenzamide | | Descriptor: | 4-{6-(cyclohexylamino)-8-[(tetrahydro-2H-pyran-4-ylmethyl)amino]imidazo[1,2-b]pyridazin-3-yl}-N-cyclopropylbenzamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
3WYX
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 6-((3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-2-(cyclohexylamino)nicotinonitrile | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
3WZK
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH N-cyclopropyl-4-(8-((thiophen-2-ylmethyl)amino)imidazo[1,2-a]pyrazin-3-yl)benzamide | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
3WYY
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH (E)-3-(4-((6-(((3s,5s,7s)-adamantan-1-yl)amino)-4-amino-5-cyanopyridin-2-yl)amino)-2-(cyanomethoxy)phenyl)-N-(2-methoxyethyl)acrylamide | | Descriptor: | (2E)-3-[4-({4-amino-5-cyano-6-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]pyridin-2-yl}amino)-2-(cyanomethoxy)phenyl]-N-(2-methoxyethyl)prop-2-enamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Murai, H, Nakamura, Y. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
6Y3H
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0401 | | Descriptor: | Major allergen Cor a 1.0401 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3L
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0404 | | Descriptor: | Major allergen variant Cor a 1.0404 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3I
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0402 | | Descriptor: | Major allergen variant Cor a 1.0402 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
6Y3K
 
 | | NMR solution structure of the hazelnut allergen Cor a 1.0403 | | Descriptor: | Major allergen variant Cor a 1.0403 | | Authors: | Fuehrer, S, Kamenik, A.S, Zeindl, R, Nothegger, B, Hofer, F, Reider, N, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Inverse relation between structural flexibility and IgE reactivity of Cor a 1 hazelnut allergens.
Sci Rep, 11, 2021
|
|
1PIM
 
 | | DITHIONITE REDUCED E. COLI RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT, D84E MUTANT | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
1PIU
 
 | | OXIDIZED RIBONUCLEOTIDE REDUCTASE R2-D84E MUTANT CONTAINING OXO-BRIDGED DIFERRIC CLUSTER | | Descriptor: | FE (III) ION, MERCURY (II) ION, OXYGEN ATOM, ... | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
7O00
 
 | | Crystal structure of HLA-DR4 in complex with a HSP70 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaperone protein DnaK, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZH
 
 | | Crystal structure of HLA-DR4 in complex with a citrullinated cilp peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZF
 
 | | Crystal structure of HLA-DR4 in complex with a mutated human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZE
 
 | | Crystal structure of HLA-DR4 in complex with a human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen alpha-1(II) chain, GLYCEROL, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
2ZDI
 
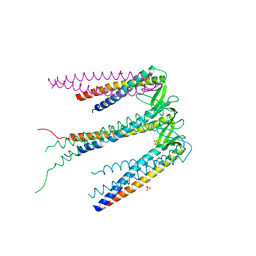 | | Crystal structure of Prefoldin from Pyrococcus horikoshii OT3 | | Descriptor: | Prefoldin subunit alpha, Prefoldin subunit beta, SULFATE ION | | Authors: | Kida, H, Miki, K. | | Deposit date: | 2007-11-23 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins
J.Mol.Biol., 376, 2008
|
|
8PM2
 
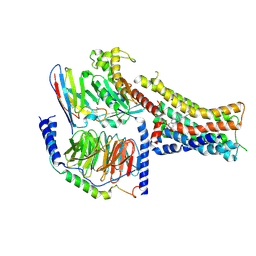 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
8F76
 
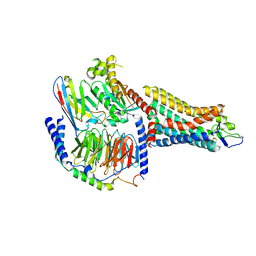 | | Human olfactory receptor OR51E2 bound to propionate in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Billesboelle, C.B, Manglik, A. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of odorant recognition by a human odorant receptor.
Nature, 615, 2023
|
|
7AD3
 
 | | Class D GPCR Ste2 dimer coupled to two G proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-factor mating pheromone, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Velazhahan, V, Tate, C. | | Deposit date: | 2020-09-14 | | Release date: | 2020-12-09 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the class D GPCR Ste2 dimer coupled to two G proteins.
Nature, 589, 2021
|
|
6XG4
 
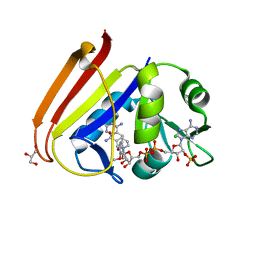 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
6XG5
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
7QBC
 
 | |
