3ZUM
 
 | | Photosynthetic Reaction Centre Mutant with Phe L146 replaced with Ala | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
3ZUW
 
 | | Photosynthetic Reaction Centre Mutant with TYR L128 replaced with HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
5NYO
 
 | | Crystal structure of an atypical poplar thioredoxin-like2.1 variant in dimeric form | | Descriptor: | SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYK
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in oxidized state | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYN
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYM
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in reduced state | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYL
 
 | |
5YGV
 
 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist | | Descriptor: | (2Z,4E)-3-methyl-5-[(1S,4S)-2,6,6-trimethyl-4-[3-(4-methylphenyl)prop-2-ynoxy]-1-oxidanyl-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Mimura, N, Okamoto, M, Monda, K, Iba, K, Ohnishi, T, Todoroki, Y, Yajima, S. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions.
ACS Chem. Biol., 13, 2018
|
|
4UBQ
 
 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
6EN5
 
 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | Descriptor: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | Authors: | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
5JR0
 
 | |
8PKJ
 
 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKI
 
 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
5GUG
 
 | | Crystal structure of inositol 1,4,5-trisphosphate receptor large cytosolic domain with inositol 1,4,5-trisphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Hamada, K, Miyatake, H, Terauchi, A, Mikoshiba, K. | | Deposit date: | 2016-08-29 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (7.399 Å) | | Cite: | IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3JAO
 
 | | Ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Maheshwari, A, Obbineni, J.M, Bui, K.H, Shibata, K, Toyoshima, Y.Y, Ishikawa, T. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | alpha- and beta-Tubulin Lattice of the Axonemal Microtubule Doublet and Binding Proteins Revealed by Single Particle Cryo-Electron Microscopy and Tomography.
Structure, 23, 2015
|
|
4BXK
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with a domain-specific inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Douglas, R.G, Sharma, R.K, Masuyer, G, Lubbe, L, Zamora, I, Acharya, K.R, Chibale, K, Sturrock, E.D. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Design for the Development of N-Domain Selective Angiotensin-1 Converting Enzyme Inhibitors
Clin.Sci., 126, 2014
|
|
5MP7
 
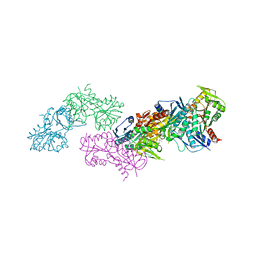 | | Crystal structure of phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Donini, S, Garavaglia, S, Ferraris, D.M, Miggiano, R, Mori, S, Shibayama, K, Rizzi, M. | | Deposit date: | 2016-12-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural investigations on phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis.
PLoS ONE, 12, 2017
|
|
4GPS
 
 | |
4GPU
 
 | |
1N4K
 
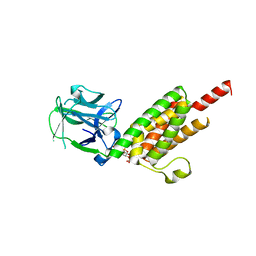 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2002-10-31 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
6KXX
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
6KXY
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound B) | | Descriptor: | 6-ethyl-1-(4-fluorophenyl)-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
