5MRW
 
 | | Structure of the KdpFABC complex | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Huang, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the potassium-importing KdpFABC membrane complex.
Nature, 546, 2017
|
|
2B4C
 
 | | Crystal structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) complexed with CD4 and the X5 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, T-cell surface glycoprotein CD4, ... | | Authors: | Huang, C, Tang, M, Zhang, M.Y, Majeed, S, Montabana, E, Stanfield, R.L, Dimitrov, D.S, Korber, B, Sodroski, J, Wilson, I.A, Wyatt, R, Kwong, P.D. | | Deposit date: | 2005-09-23 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a V3-containing HIV-1 gp120 core.
Science, 310, 2005
|
|
7BZH
 
 | |
2RD0
 
 | | Structure of a human p110alpha/p85alpha complex | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations.
Science, 318, 2007
|
|
5YJO
 
 | |
6IJL
 
 | |
8W5J
 
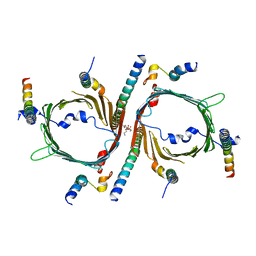 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
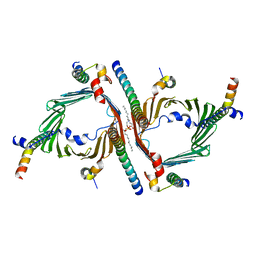 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
5S9R
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMS-986158, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9P
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 9-benzyl-2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9H-carbazole-4-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-benzyl-2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9H-carbazole-4-carboxamide, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9O
 
 | | CRYSTAL STRUCTURE OF THE HUMAN BRD2 BD1 BROMODOMAIN IN COMPLEX WITH 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide, Bromodomain-containing protein 2 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9Q
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9-[(S)-(oxan-4-yl)(phenyl)methyl]-9H-carbazole-4-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9-[(S)-(oxan-4-yl)(phenyl)methyl]-9H-carbazole-4-carboxamide, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
6NZH
 
 | |
6NZF
 
 | |
6NZE
 
 | |
7UZN
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMT-206059 AKA 2-{(3M)-3-(1,4-DIMETHYL-1H-1,2,3-TRIAZOL-5-YL)-8-FLUORO-5-[(S)-(OXAN-4-YL)(PHENYL)METHYL]-5H-PYRIDO[3,2-b]INDOL-7-YL}PROPAN-2-OL, TRIPLY DEUTERATED ON THE 4-METHYL GROUP | | Descriptor: | 1,2-ETHANEDIOL, 2-{(3M)-3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Development of BET Inhibitors as Potential Treatments for Cancer: Optimization of Pharmacokinetic Properties.
Acs Med.Chem.Lett., 13, 2022
|
|
6NZR
 
 | |
6NZQ
 
 | |
6NZP
 
 | |
7ROV
 
 | | KRAS G12D Mutant in complex with GMPPCP and cyclic peptide MP-9903 | | Descriptor: | Cyclic peptide MP-9903, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Orth, P. | | Deposit date: | 2021-08-02 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of cell active macrocyclic peptides with on-target inhibition of KRAS signaling.
Chem Sci, 12, 2021
|
|
1L3M
 
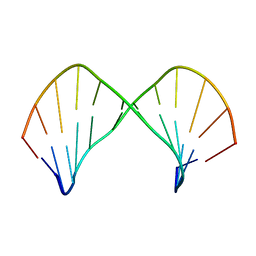 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
7MP5
 
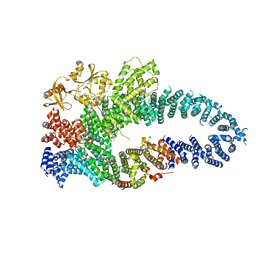 | | Autoinhibited neurofibrobmin | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOC
 
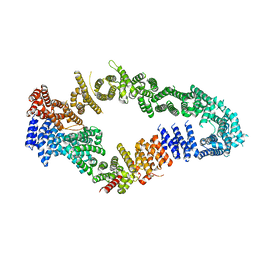 | | Neurofibromin core | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-15 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MP6
 
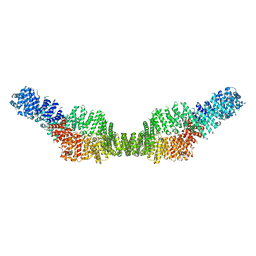 | | Neurofibromin homodimer | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5TKB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 4D IN COMPLEX WITH A TETRAFLUORANLINE COMPOUND | | Descriptor: | ETHANOL, MAGNESIUM ION, N-[(2R)-2,3-dihydroxy-2-methylpropyl]-8-(methylamino)-6-[(2,3,5,6-tetrafluorophenyl)amino]imidazo[1,2-b]pyridazine-3-carboxamide, ... | | Authors: | Sack, J.S. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
