1L8D
 
 | | Rad50 coiled-coil Zn hook | | Descriptor: | CITRIC ACID, DNA double-strand break repair rad50 ATPase, MERCURY (II) ION, ... | | Authors: | Hopfner, K.P, Tainer, J.A. | | Deposit date: | 2002-03-20 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair.
Nature, 418, 2002
|
|
1F2T
 
 | | Crystal Structure of ATP-Free RAD50 ABC-ATPase | | Descriptor: | RAD50 ABC-ATPASE | | Authors: | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
1F2U
 
 | | Crystal Structure of RAD50 ABC-ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAD50 ABC-ATPASE | | Authors: | Hopfner, K.P, Karcher, A, Shin, D.S, Craig, L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily.
Cell(Cambridge,Mass.), 101, 2000
|
|
1FXY
 
 | | COAGULATION FACTOR XA-TRYPSIN CHIMERA INHIBITED WITH D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | COAGULATION FACTOR XA-TRYPSIN CHIMERA, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | Authors: | Hopfner, K.P, Kopetzki, E, Kresse, G.-B, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | New enzyme lineages by subdomain shuffling.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1S8E
 
 | | Crystal structure of Mre11-3 | | Descriptor: | MANGANESE (II) ION, exonuclease putative | | Authors: | Hopfner, K.P. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Mre11-3
Nucleic Acids Res., 32, 2004
|
|
2BA1
 
 | | Archaeal exosome core | | Descriptor: | Archaeal exosome RNA binding protein CSL4, Archaeal exosome complex exonuclease RRP41, Archaeal exosome complex exonuclease RRP42, ... | | Authors: | Hopfner, K.P, Buttner, K, Wenig, K. | | Deposit date: | 2005-10-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural framework for the mechanism of archaeal exosomes in RNA processing.
Mol.Cell, 20, 2005
|
|
3RA7
 
 | |
4LEO
 
 | |
8ATF
 
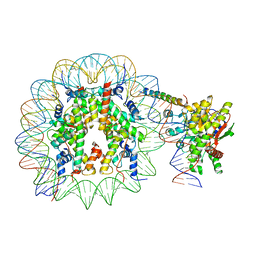 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8AV6
 
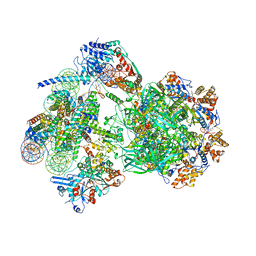 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8BAH
 
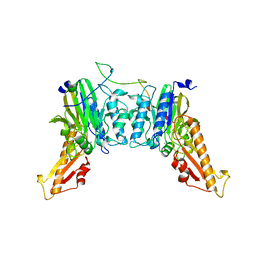 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
2W4R
 
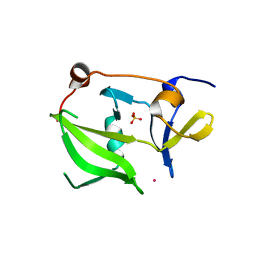 | | Crystal structure of the regulatory domain of human LGP2 | | Descriptor: | MERCURY (II) ION, PROBABLE ATP-DEPENDENT RNA HELICASE DHX58, SULFATE ION | | Authors: | Pippig, D.A, Hellmuth, J.C, Cui, S, Kirchhofer, A, Lammens, K, Lammens, A, Schmidt, A, Rothenfusser, S, Hopfner, K.P. | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Regulatory Domain of the Rig-I Family ATPase Lgp2 Senses Double-Stranded RNA.
Nucleic Acids Res., 37, 2009
|
|
2XGP
 
 | | Yeast DNA polymerase eta in complex with C8-2-acetylaminofluorene containing DNA | | Descriptor: | 5'-D(*CP*8FG*CP*TP*CP*AP*TP*CP*CP*AP*C)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*AP*G)-3', CALCIUM ION, ... | | Authors: | Scheider, S, Lammens, K, Schorr, S, Hopfner, K.P, Carell, T. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Replication Blocking and Bypass of Y-Family Polymerase Eta by Bulky Acetylaminofluorene DNA Adducts.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5M1X
 
 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
4ON9
 
 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1US8
 
 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
6GK2
 
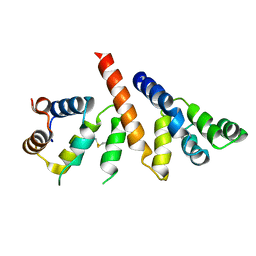 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
3C1Y
 
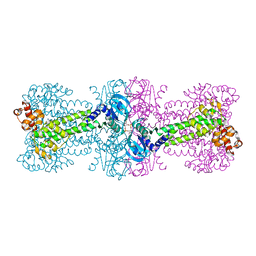 | | Structure of bacterial DNA damage sensor protein with co-purified and co-crystallized ligand | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3C21
 
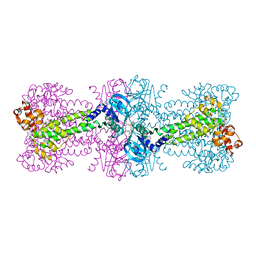 | | Structure of a bacterial DNA damage sensor protein with reaction product | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3C23
 
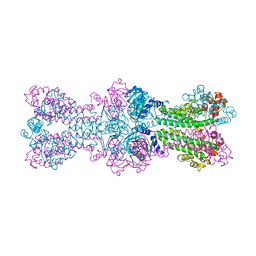 | | Structure of a bacterial DNA damage sensor protein with non-reactive Ligand | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA integrity scanning protein disA | | Authors: | Witte, G, Hartung, S, Buttner, K, Hopfner, K.P. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates
Mol.Cell, 30, 2008
|
|
3BK7
 
 | |
3C1Z
 
 | |
5NBM
 
 | | Crystal structure of the Arp4-N-actin(ATP-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5NBN
 
 | | Crystal structure of the Arp4-N-actin-Arp8-Ino80HSA module of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5NBL
 
 | | Crystal structure of the Arp4-N-actin(APO-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
