6W4I
 
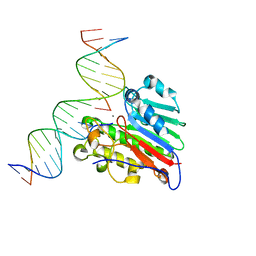 | | APE1 Y269A product complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
6W4T
 
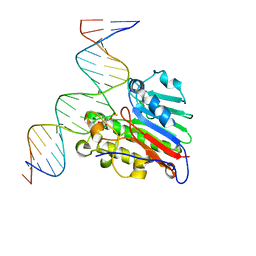 | | APE1 Y269A phosphorothioate substrate complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
7TR7
 
 | | APE1 product complex with abasic ssDNA | | Descriptor: | DNA (5'-D(P*(3DR)P*CP*GP*AP*TP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2022-01-28 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into AP-endonuclease 1 cleavage of abasic sites at stalled replication fork mimics.
Nucleic Acids Res., 51, 2023
|
|
9DP3
 
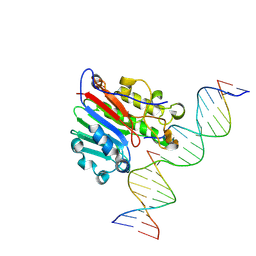 | | APE1 N174D Product Complex with Abasic DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Hoitsma, N.M, DeHart, K.D, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N174 Physically and Chemically Stabilizes Abasic DNA During the APE1 Endonuclease Reaction
To Be Published
|
|
7LPH
 
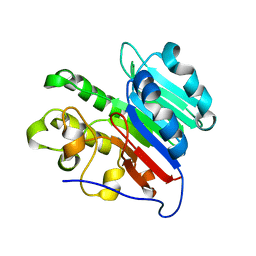 | | APE1 Mn-bound product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPJ
 
 | | APE1 Mn-bound phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPG
 
 | | APE1 product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPI
 
 | | APE1 phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
8THW
 
 | | Cac1 PIP motif bound to PCNA | | Descriptor: | Proliferating cell nuclear antigen,Chromatin assembly factor 1 subunit p90 | | Authors: | Veltri, E, Hoitsma, N.M, Dieckman, L. | | Deposit date: | 2023-07-18 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Interaction Between Yeast Chromatin Assembly Factor 1 and Proliferating Cell Nuclear Antigen.
J.Mol.Biol., 436, 2024
|
|
8UA7
 
 | | Medusavirus Nucleosome Core Particle | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Toner, C.M, Hoitsma, N.M, Weerawarana, S, Luger, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of Medusavirus encoded histones reveals nucleosome-like structures and a unique linker histone.
Nat Commun, 15, 2024
|
|
9DP4
 
 | | APE1 N174Q Product Complex with Abasic DNA | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | DeHart, K.D, Hoitsma, N.M, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | N174 Physically and Chemically Stabilizes the APE1 Endonuclease Reaction
To Be Published
|
|
6UOL
 
 | |
6UOK
 
 | |
6UOM
 
 | | Y271G DNA polymerase beta ternary complex with templating adenine and incoming r8-oxo-GTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, M.R, Freudenthal, B.D. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and structural characterization of oxidized ribonucleotide insertion into DNA by human DNA polymerase beta.
J.Biol.Chem., 295, 2020
|
|
7U51
 
 | |
7U50
 
 | |
7U52
 
 | |
7U53
 
 | |
9DP2
 
 | | APE1 N174A Product Complex with Abasic DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | DeHart, K.D, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | N174 Physically and Chemically Stabilizes Abasic DNA During the APE1 Endonuclease Reaction
To Be Published
|
|
9DP1
 
 | | APE1 N174A Substrate Complex with Abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA repair nuclease/redox regulator APEX1, ... | | Authors: | DeHart, K.M, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N174 Physically and Chemically Stabilizes the Endonuclease Reaction of APE1
To Be Published
|
|
