6GP2
 
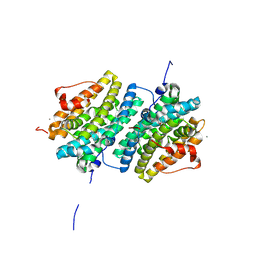 | | Ribonucleotide Reductase class Ie R2 from Mesoplasma florum, DOPA-active form | | Descriptor: | CALCIUM ION, Ribonucleoside-diphosphate reductase beta chain | | Authors: | Srinivas, V, Lebrette, H, Lundin, D, Kutin, Y, Sahlin, M, Lerche, M, Enrich, J, Branca, R.M.M, Cox, N, Sjoberg, B.M, Hogbom, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens.
Nature, 563, 2018
|
|
5OC0
 
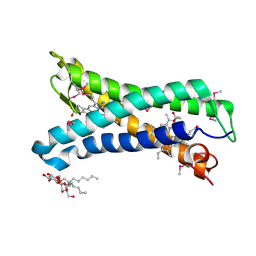 | | Structure of E. coli superoxide oxidase | | Descriptor: | Cytochrome b561, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lundgren, C.A.K, Sjostrand, D, Biner, O, Bennett, M, von Ballmoos, C, Hogbom, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-06-20 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Scavenging of superoxide by a membrane-bound superoxide oxidase.
Nat. Chem. Biol., 14, 2018
|
|
6I90
 
 | |
6I93
 
 | |
6I92
 
 | |
6R81
 
 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6I95
 
 | |
6I94
 
 | |
6F6M
 
 | |
6F6L
 
 | |
6F6B
 
 | |
6F65
 
 | |
2WOC
 
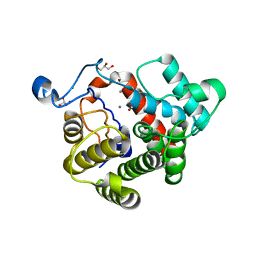 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOE
 
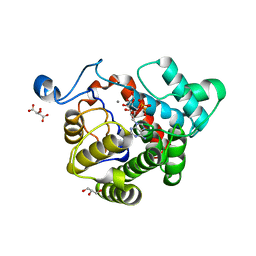 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOD
 
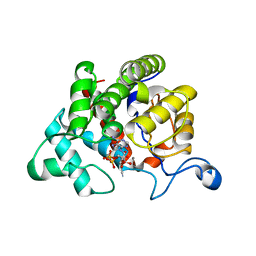 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XCJ
 
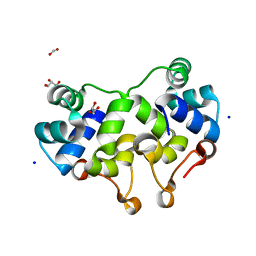 | | Crystal structure of P2 C, the immunity repressor of temperate E. coli phage P2 | | Descriptor: | C PROTEIN, FORMIC ACID, GLYCEROL, ... | | Authors: | Massad, T, Skaar, K, Hogbom, M, Stenmark, P. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the P2 C-Repressor: A Binder of Non-Palindromic Direct DNA Repeats.
Nucleic Acids Res., 38, 2010
|
|
2FIM
 
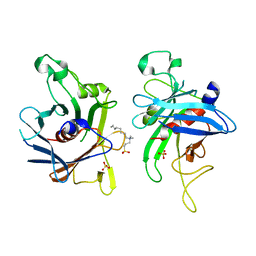 | | Structure of the C-terminal domain of Human Tubby-like protein 1 | | Descriptor: | 3-(N,N-DIMETHYLOCTYLAMMONIO)PROPANESULFONATE, SULFATE ION, Tubby related protein 1 | | Authors: | Hallberg, B.M, Ogg, D, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Persson, C. | | Deposit date: | 2005-12-29 | | Release date: | 2006-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Human Tubby-like protein 1
To be published
|
|
5NF8
 
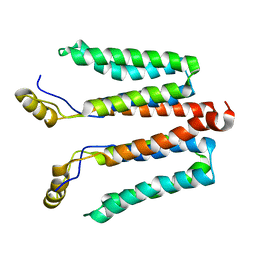 | | Solution structure of detergent-solubilized Rcf1, a yeast mitochondrial inner membrane protein involved in respiratory Complex III/IV supercomplex formation | | Descriptor: | Respiratory supercomplex factor 1, mitochondrial | | Authors: | Zhou, S, Pettersson, P, Sjoholm, J, Sjostrand, D, Hogbom, M, Brzezinski, P, Maler, L, Adelroth, P. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast Rcf1, a protein involved in respiratory supercomplex formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OCV
 
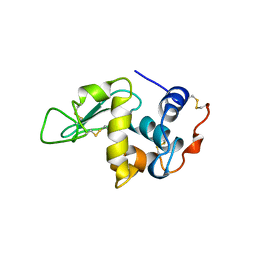 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
5OVO
 
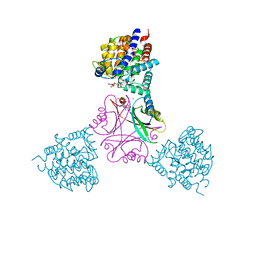 | |
5OMK
 
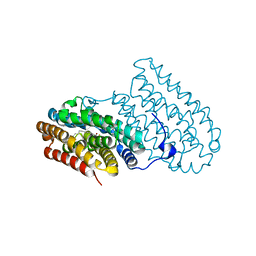 | |
5OMJ
 
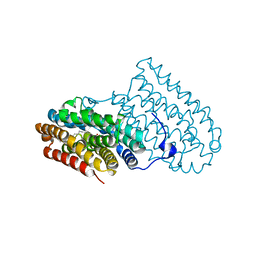 | |
2H4U
 
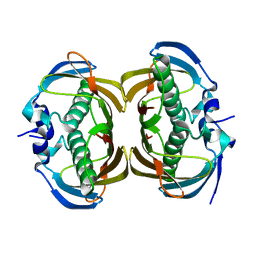 | | Crystal Structure of Human Thioesterase Superfamily Member 2 | | Descriptor: | Thioesterase superfamily member 2 | | Authors: | Ogg, D.J, Uppenberg, J, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Grasslund, S, Flodin, S, Hammerstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.-G, Weigelt, J, Hallberg, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human thioesterase superfamily member 2
To be Published
|
|
2J91
 
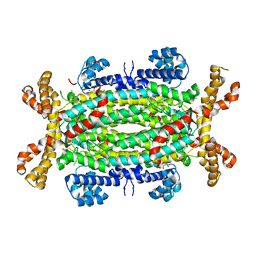 | | Crystal structure of Human Adenylosuccinate Lyase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, CHLORIDE ION, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Adenylosuccinate Lyase
To be Published
|
|
6H5A
 
 | |
