1KTZ
 
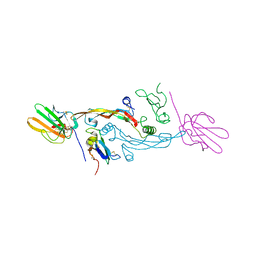
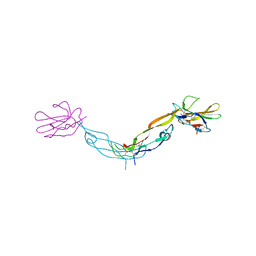 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
7SXB
 
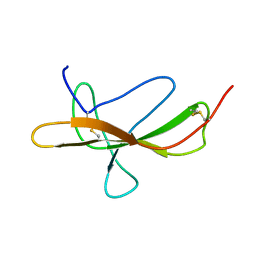 | |
2PJY
 
 | |
2JQE
 
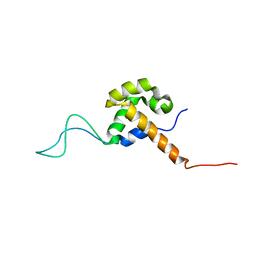 | |
8G4K
 
 | |
6MZP
 
 | |
6MZN
 
 | |
2L5S
 
 | | Solution structure of the extracellular domain of the TGF-beta type I receptor | | Descriptor: | TGF-beta receptor type-1 | | Authors: | Zuniga, J.E, Ilangovan, U, Pardeep, M, Hinck, C, Huang, T. | | Deposit date: | 2010-11-04 | | Release date: | 2011-10-26 | | Method: | SOLUTION NMR | | Cite: | The TbetaR-I Pre-Helix Extension Is Structurally Ordered in the Unbound Form and Its Flanking Prolines Are Essential for Binding
J.Mol.Biol., 412, 2011
|
|
5TX4
 
 | | Derivative of mouse TGF-beta2, with a deletion of residues 52-71 and K25R, R26K, L51R, A74K, C77S, L89V, I92V, K94R T95K, I98V single amino acid substitutions, bound to human TGF-beta type II receptor ectodomain residues 15-130 | | Descriptor: | TGF-beta receptor type-2, Transforming growth factor beta-2 | | Authors: | Hinck, A.P, Kim, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
5TX6
 
 | |
5TX2
 
 | | Miniature TGF-beta2 3-mutant monomer | | Descriptor: | Transforming growth factor beta-2 | | Authors: | Taylor, A.B, Kim, S.K, Hart, P.J, Hinck, A.P. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | Descriptor: | 1,2-ETHANEDIOL, CTLA-4 binder | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
8GAD
 
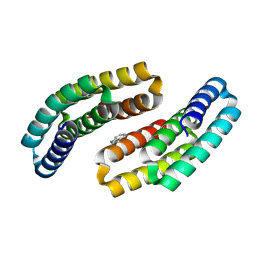 | |
8GAB
 
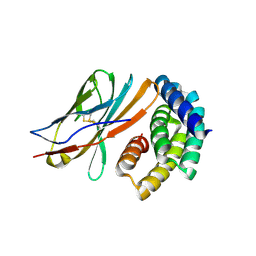 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | Descriptor: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
6MAC
 
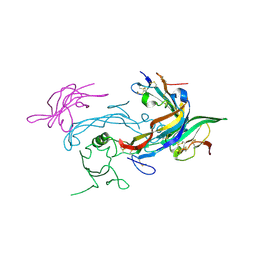 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5TY4
 
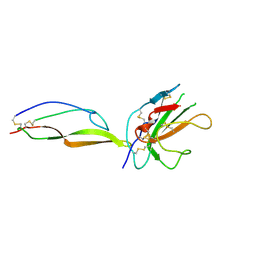 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7R
 
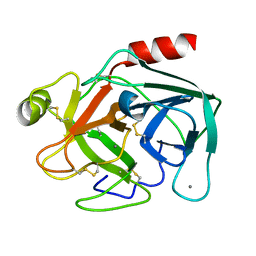 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7Q
 
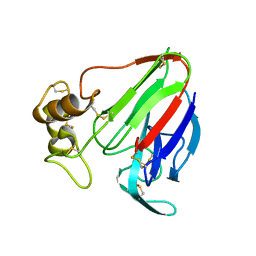 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7O
 
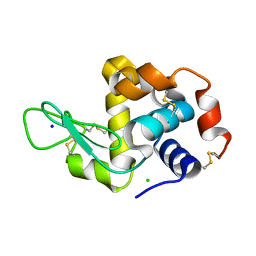 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
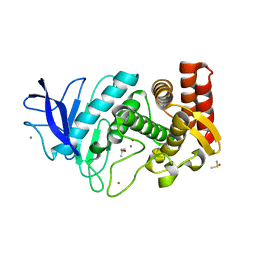 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7S
 
 | | MicroED structure of proteinase K at 1.6 A resolution | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7P
 
 | | MicroED structure of xylanase at 2.3 A resolution | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
