6W0P
 
 | | Putative kojibiose phosphorylase from human microbiome | | Descriptor: | Kojibiose phosphorylase | | Authors: | Dementiev, A, Osipiuk, J, Endres, M, Wakatsuki, S, Hess, M, Joachimiak, A. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Putative kojibiose phosphorylase from human microbiome
to be published
|
|
8ROQ
 
 | | FAdV-C4 Aviadenovirus structure, strain KR5 | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Perez-Illana, M.P, Schachnner, A, Condezo, G.N, Hernando-Perez, M, Martinez, M, Marabini, R, Hess, M, San Martin, C. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | FAdV-C4 AG234 Aviadenovirus structure, strain KR5
To Be Published
|
|
6BBE
 
 | | Structure of N-glycosylated porcine surfactant protein-D | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
6BBD
 
 | | Structure of N-glycosylated porcine surfactant protein-D complexed with glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
4N1J
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
4N1K
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1L
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EWI
 
 | | Crystal structure of the NLRP4 Pyrin domain | | Descriptor: | CHLORIDE ION, NACHT, LRR and PYD domains-containing protein 4, ... | | Authors: | Eibl, C, Hessenberger, M, Puehringer, S, Page, R, Diederichs, K, Peti, W. | | Deposit date: | 2012-04-27 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Functional Analysis of the NLRP4 Pyrin Domain.
Biochemistry, 51, 2012
|
|
4H1V
 
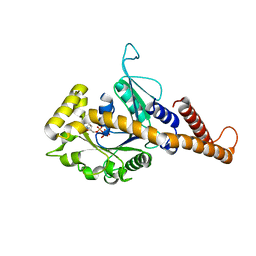 | | GMP-PNP bound dynamin-1-like protein GTPase-GED fusion | | Descriptor: | Dynamin-1-like protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Wenger, J, Klinglmayr, E, Eibl, C, Hessenberger, M, Goettig, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Mapping of Human Dynamin-1-Like GTPase Domain Based on X-ray Structure Analyses.
Plos One, 8, 2013
|
|
2XT0
 
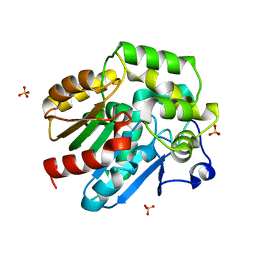 | | Dehalogenase DPpA from Plesiocystis pacifica SIR-I | | Descriptor: | HALOALKANE DEHALOGENASE, SULFATE ION | | Authors: | Bogdanovic, X, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-10-02 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cloning, Functional Expression, Biochemical Characterization, and Structural Analysis of a Haloalkane Dehalogenase from Plesiocystis Pacifica Sir-1.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
5WH8
 
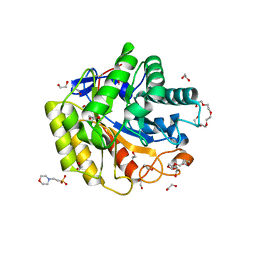 | | Cellulase Cel5C_n | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Koropatkin, N.M, Pope, P.B, Naas, A.E. | | Deposit date: | 2017-07-15 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | "Candidatus Paraporphyromonas polyenzymogenes" encodes multi-modular cellulases linked to the type IX secretion system.
Microbiome, 6, 2018
|
|
2IUM
 
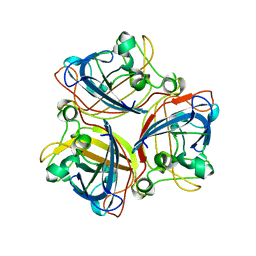 | |
2IUN
 
 | | Structure of the C-terminal head domain of the avian adenovirus CELO long fibre (P21 crystal form) | | Descriptor: | AVIAN ADENOVIRUS CELO LONG FIBRE, CALCIUM ION | | Authors: | Guardado-Calvo, P, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2006-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the C-terminal head domain of the fowl adenovirus type 1 long fiber.
J. Gen. Virol., 88, 2007
|
|
9C80
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent inhibitor | | Descriptor: | (5R,7S,8R)-7-(2-fluorophenyl)-3-[(2-fluorophenyl)carbamoyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-5-carboxylic acid, 3C-like proteinase nsp5 | | Authors: | Ornelas, E, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 67, 2024
|
|
9C7W
 
 | | human OC43 Main Protease (1-303) in complex with potent inhibitor | | Descriptor: | (8S)-3-(4,4-difluorocyclohexyl)-5-(pyrimidin-2-yl)pyrazolo[1,5-a]pyrimidine, ORF1ab polyprotein | | Authors: | Tang, J.Y, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 67, 2024
|
|
9C8Q
 
 | |
4GV7
 
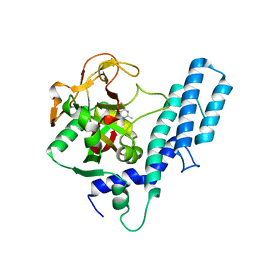 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor ME0328 | | Descriptor: | 2-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
4GV2
 
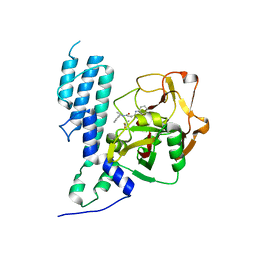 | | Human ARTD3 (PARP3) - Catalytic domain in complex with inhibitor ME0354 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1R)-1-(pyridin-2-yl)ethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
4GV0
 
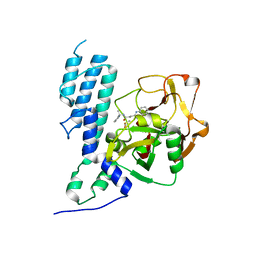 | | Human ARTD3 (PARP3) - Catalytic domain in complex with inhibitor ME0355 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-(pyridin-2-yl)ethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
4GV4
 
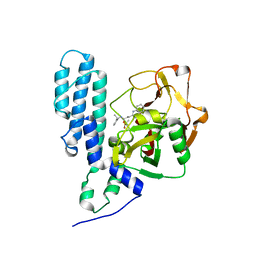 | | Human ARTD3 (PARP3) - Catalytic domain in complex with inhibitor ME0328 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-phenylethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
4MOY
 
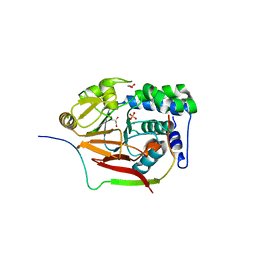 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
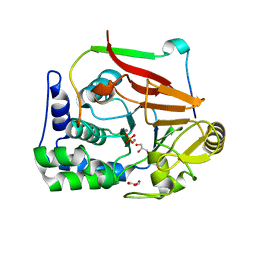 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MOV
 
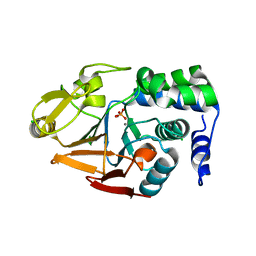 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6RZU
 
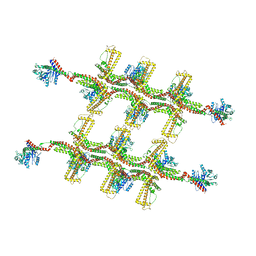 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
