3D8T
 
 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | ACETATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8S
 
 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | GLYCEROL, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8N
 
 | | Uroporphyrinogen III Synthase-Uroporphyringen III Complex | | Descriptor: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8R
 
 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | PHOSPHATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
5UMU
 
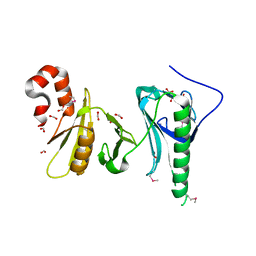 | | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16 | | Descriptor: | ACETATE ION, FACT complex subunit SPT16, FORMIC ACID | | Authors: | Hu, Q, Thompson, J.R, Heroux, A, Su, D, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the middle double PH domain of human FACT complex subunit SPT16
To Be Published
|
|
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
3LZU
 
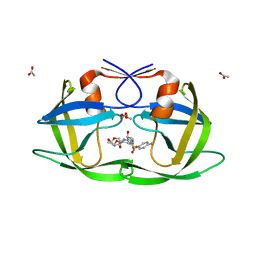 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3LZV
 
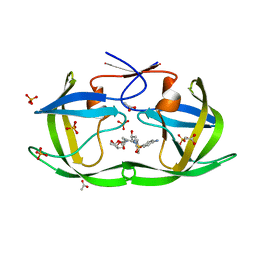 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3NPQ
 
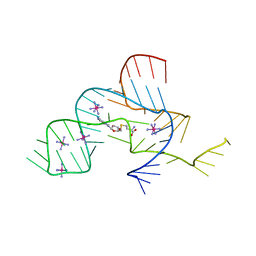 | |
3NPN
 
 | |
3OSS
 
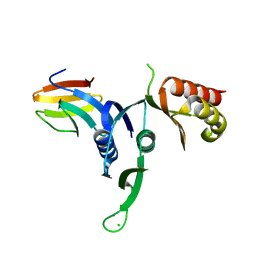 | | The crystal structure of enterotoxigenic Escherichia coli GspC-GspD complex from the type II secretion system | | Descriptor: | CALCIUM ION, CHLORIDE ION, TYPE 2 SECRETION SYSTEM, ... | | Authors: | Korotkov, K.V, Pruneda, J, Hol, W.G.J. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and functional studies on the interaction of GspC and GspD in the type II secretion system.
Plos Pathog., 7, 2011
|
|
3PL6
 
 | |
1OMO
 
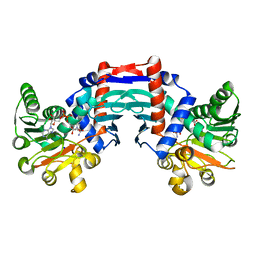 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
4EVO
 
 | |
4DFA
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DGZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DF7
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23L/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DU9
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/V74A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
3RCP
 
 | | Crystal structure of the FAPP1 pleckstrin homology domain | | Descriptor: | GLYCEROL, Pleckstrin homology domain-containing family A member 3 | | Authors: | Roy, S, He, J, Kutateladze, T.G. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Phosphatidylinositol 4-Phosphate and ARF1 GTPase Recognition by the FAPP1 Pleckstrin Homology (PH) Domain.
J.Biol.Chem., 286, 2011
|
|
3SZM
 
 | |
3THF
 
 | |
3V46
 
 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
3T16
 
 | |
3TME
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+NVIAGLA V23E at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+NVIAGLA V23E at cryogenic temperature
To be Published
|
|
