1D7Z
 
 | |
7TQW
 
 | | Kod RSGA incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
4Y8D
 
 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
8SV4
 
 | | 7-Deazapurines and 5-Halogenpyrimidine DNA duplex | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, ACETATE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Morphing by a DNA Analogue Featuring 7-Deazapurines and 5-Halogenpyrimidines and the Origins of Adenine-Tract Geometry.
Biochemistry, 62, 2023
|
|
8SV3
 
 | | 7-Deazapurines and 5-Halogenpyrimidine DNA duplex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Modified DNA, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Conformational Morphing by a DNA Analogue Featuring 7-Deazapurines and 5-Halogenpyrimidines and the Origins of Adenine-Tract Geometry.
Biochemistry, 62, 2023
|
|
5L4Q
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase 1 (AAK1) in Complex with LKB1 (AAK1 Dual Inhibitor) | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, ~{N}-[5-(4-cyanophenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]pyridine-3-carboxamide | | Authors: | Sorrell, F.J, Williams, E, Fox, N, Abdul Azeez, K.R, Gileadi, O, von Delft, F, Edwards, A.M, Bountra, C, Elkins, J.M, Knapp, S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 3,5-Disubstituted-pyrrolo[2,3- b]pyridines as Inhibitors of Adaptor-Associated Kinase 1 with Antiviral Activity.
J.Med.Chem., 2019
|
|
3OK2
 
 | |
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
4HTU
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUG
 
 | | Structure of 5-chlorouracil modified A:U base pairs | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUF
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*(UCL)P*TP*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
4HUE
 
 | | Structure of 5-chlorouracil modified G:U base pair | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(UCL)P*GP*CP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
3KNC
 
 | |
6TBC
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin B | | Descriptor: | (2~{E},5~{R},10~{E},12~{E},15~{S},19~{R})-20-[[(2~{R},3~{R})-3-aminocarbonyloxy-2-methyl-butanoyl]amino]-3,5,15-trimethyl-7-methylidene-19-oxidanyl-17-oxidanylidene-icosa-2,10,12-trienoic acid, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6TBB
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and kalimantacin A (batumin) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], Kalimantacin, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fage, C.D, Masschelein, J. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1KI2
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI3
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH PENCICLOVIR | | Descriptor: | 9-(4-HYDROXY-3-(HYDROXYMETHYL)BUT-1-YL)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Jarvest, R.L, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI7
 
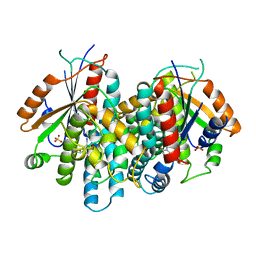 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-IODODEOXYURIDINE | | Descriptor: | 5-IODODEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KIM
 
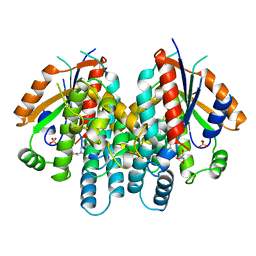 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH DEOXYTHYMIDINE | | Descriptor: | SULFATE ION, THYMIDINE, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI8
 
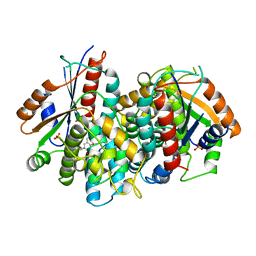 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOVINYLDEOXYURIDINE | | Descriptor: | 5-BROMOVINYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1KI4
 
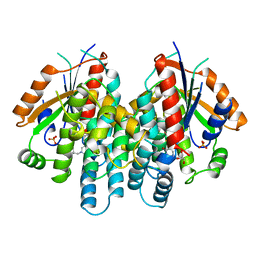 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOTHIENYLDEOXYURIDINE | | Descriptor: | 5-BROMOTHIENYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
7OTK
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-233 | | Descriptor: | (2~{R})-2-[2-(6-aminopurin-9-yl)ethylamino]-3-phosphono-propanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTN
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | Descriptor: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND inhibitor RMC-282 | | Descriptor: | (R)-N-(1-(6-amino-9H-purin-9-yl)propan-2-yl)-N-(2-phosphonoethyl)glycine, DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTA
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | Descriptor: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
