3JS1
 
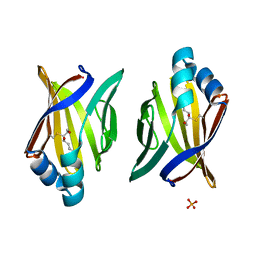 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3JSQ
 
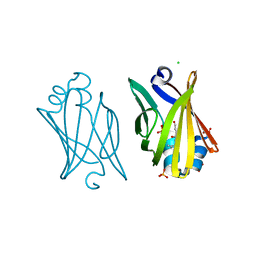 | | Crystal structure of adipocyte fatty acid binding protein non-covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, Adipocyte fatty acid-binding protein, CHLORIDE ION, ... | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
2GUZ
 
 | | Structure of the Tim14-Tim16 complex of the mitochondrial protein import motor | | Descriptor: | CITRATE ANION, Mitochondrial import inner membrane translocase subunit TIM14, Mitochondrial import inner membrane translocase subunit TIM16 | | Authors: | Mokranjac, D, Bourenkov, G, Hell, K, Neupert, W, Groll, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor.
Embo J., 25, 2006
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
6UAP
 
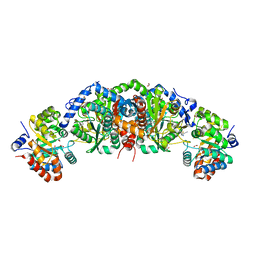 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound
To be Published
|
|
6UB9
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM
To be Published
|
|
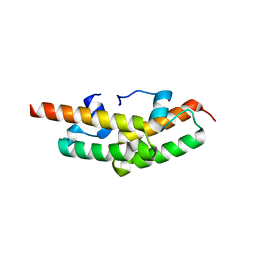
2PYR
 
 | | PHOTOACTIVE YELLOW PROTEIN, 1 NANOSECOND INTERMEDIATE (287K) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Perman, B, Srajer, V, Ren, Z, Teng, T.Y, Pradervand, C, Ursby, T, Bourgeois, D, Schotte, F, Wulff, M, Kort, R, Hellingwerf, K, Moffat, K. | | Deposit date: | 1998-03-04 | | Release date: | 1999-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energy transduction on the nanosecond time scale: early structural events in a xanthopsin photocycle.
Science, 279, 1998
|
|
6DWE
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and BRD0059-bound form | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-26 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
7OO6
 
 | | Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel of small conductance (MscS) | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OOA
 
 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO0
 
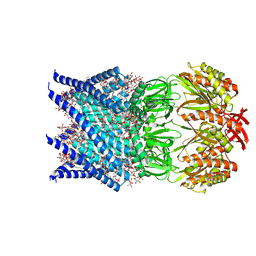 | | Mechanosensitive channel MscS solubilized with DDM in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Mechanosensitive channel MscS | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OO8
 
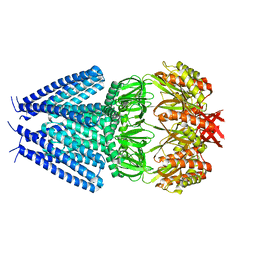 | |
7ONJ
 
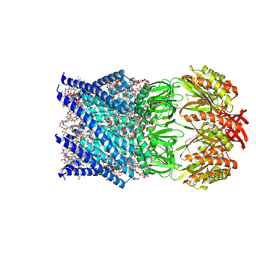 | | Mechanosensitive channel MscS solubilized with LMNG in open conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Rasmussen, T, Flegler, V.J, Boettcher, B. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanosensitive channel gating by delipidation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ONL
 
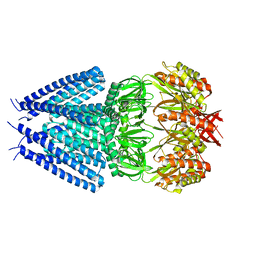 | |
6NA4
 
 | | Co crystal structure of ECR with Butryl-CoA | | Descriptor: | 9-ETHYL-9H-PURIN-6-YLAMINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA3
 
 | | Crystal Structure of Apo-form of ECR | | Descriptor: | CHLORIDE ION, Putative crotonyl-CoA reductase, Pyrrolidine | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA6
 
 | |
6NA5
 
 | | Crystal Structure of ECR in complex with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative crotonyl-CoA reductase | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
8OKH
 
 | |
4EQQ
 
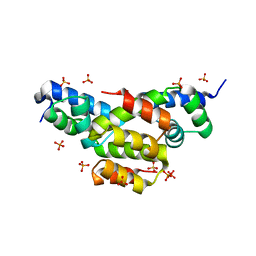 | | Structure of Ltp, a superinfection exclusion protein from the Streptococcus thermophilus temperate phage TP-J34 | | Descriptor: | PHOSPHATE ION, Putative host cell surface-exposed lipoprotein, SULFATE ION | | Authors: | Bebeacua, C, Lorenzo, C, Blangy, S, Spinelli, S, Heller, K, Cambillau, C. | | Deposit date: | 2012-04-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target.
Mol.Microbiol., 89, 2013
|
|
7BAT
 
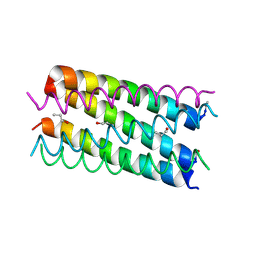 | | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(GgIaId)4 | | Descriptor: | CC-Type2-(GgIaId)4, N-PROPANOL | | Authors: | Martin, F.J.O, Dawson, W.M, Shelley, K, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-12-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
7BAS
 
 | | A de novo pentameric coiled-coil assembly: CC-Type2-(TgLaId)4-W19BrPhe. | | Descriptor: | CC-Type2-(TgLaId)4-W19BrPhe. | | Authors: | Martin, F.J.O, Dawson, W.M, Shelley, K, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-12-16 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
7BAU
 
 | | A de novo pentameric coiled-coil assembly: CC-Type2-(TgIaId)4-W19BrPhe. | | Descriptor: | CC-Type2-(TgLaId)4-W19BrPhe. | | Authors: | Martin, F.J.O, Dawson, W.M, Shelley, K, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2020-12-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
