9BZG
 
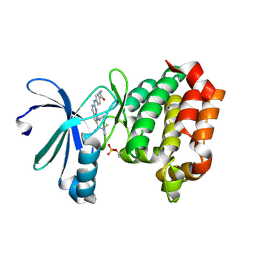 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-[4-(methylcarbamoyl)anilino]-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
9BZL
 
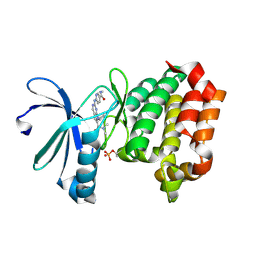 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(methylcarbamoyl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
6WGX
 
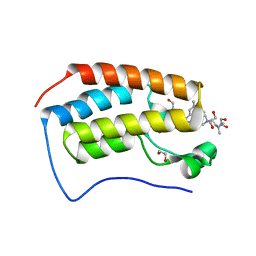 | | Cocrystal of BRD4(D1) with a selective inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7RXS
 
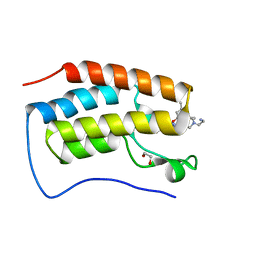 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
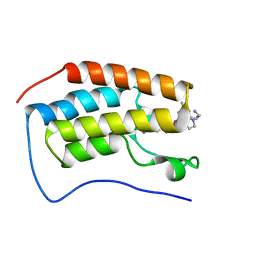 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
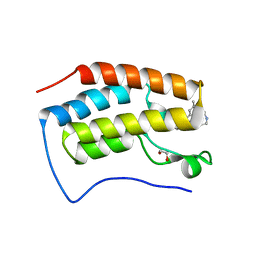 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7R9C
 
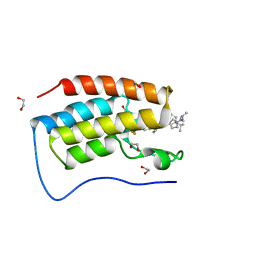 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
3BSN
 
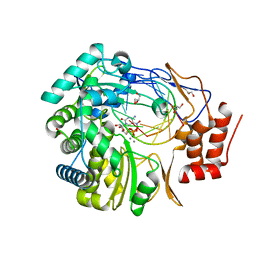 | |
3BSO
 
 | |
8FOW
 
 | | Ternary complex of CDK2 with small molecule ligands TW8672 and Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP5
 
 | | CDK2 liganded with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclin-dependent kinase 2, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP0
 
 | |
8TFD
 
 | | Crystal structure of a stem-loop DNA aptamer complexed with SARS-CoV-2 nucleocapsid protein RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DNA aptamer, Nucleoprotein | | Authors: | Esler, M, Belica, C, Shi, K, Aihara, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain.
Nucleic Acids Res., 52, 2024
|
|
3V4K
 
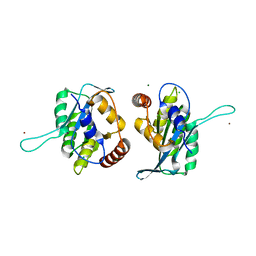 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | CHLORIDE ION, DNA dC->dU-editing enzyme APOBEC-3G, MAGNESIUM ION, ... | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
3V4J
 
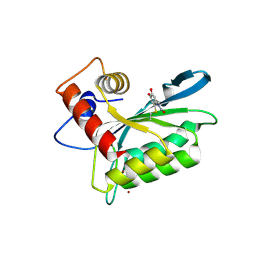 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | 4-[methyl(nitroso)amino]benzene-1,2-diol, DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
5TD5
 
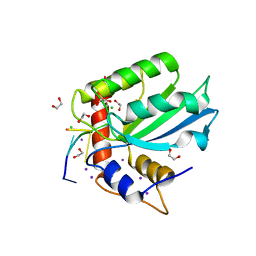 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5SWW
 
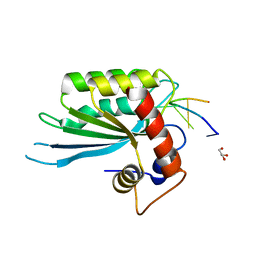 | | Crystal Structure of Human APOBEC3A complexed with ssDNA | | Descriptor: | DNA 15-Mer, DNA dC->dU-editing enzyme APOBEC-3A, GLYCEROL, ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6MH7
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SKT-68, a 1,4,5-trisubstituted imidazole analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2018-09-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase-Bromodomain Inhibitor.
J.Med.Chem., 61, 2018
|
|
6MH1
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH HU-10, A 1,4,5-Trisubstituted Imidazole Analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-(3,5-dimethylphenyl)-4-[4-(4-fluorophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2018-09-17 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase-Bromodomain Inhibitor.
J.Med.Chem., 61, 2018
|
|
6NFM
 
 | |
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
7RWF
 
 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S84
 
 | | Crystal structure of CDK2 liganded with compound TW8972 | | Descriptor: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7TGR
 
 | | Structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Esler, M.A, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2022-01-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Gain-of-Signal Assays for Probing Inhibition of SARS-CoV-2 M pro /3CL pro in Living Cells.
Mbio, 13, 2022
|
|
