2ATP
 
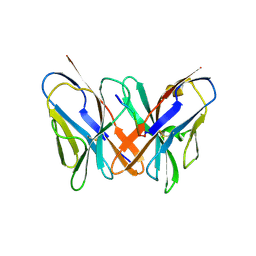 | | Crystal structure of a CD8ab heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD8 alpha chain, T-cell surface glycoprotein CD8 beta chain, ... | | Authors: | Chang, H.C, Tan, K, Ouyang, J, Parisini, E, Liu, J.H, Le, Y, Wang, X, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2005-08-25 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of a CD8alphabeta Heterodimer and Comparison with the CD8alphaalpha Homodimer.
Immunity, 23, 2005
|
|
2OW3
 
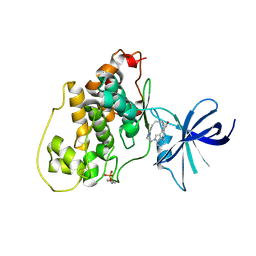 | | Glycogen synthase kinase-3 beta in complex with bis-(indole)maleimide pyridinophane inhibitor | | Descriptor: | BIS-(INDOLE)MALEIMIDE PYRIDINOPHANE, Glycogen synthase kinase-3 beta | | Authors: | Zhang, H.C, Bonaga, L.V, Ye, H, Derian, C.K, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2007-02-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6B8C
 
 | |
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
1XQS
 
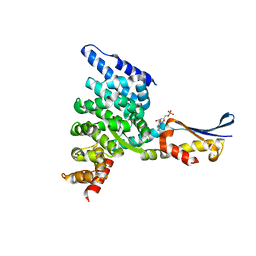 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1XQR
 
 | | Crystal structure of the HspBP1 core domain | | Descriptor: | HspBP1 protein | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1BQH
 
 | | MURINE CD8AA ECTODOMAIN FRAGMENT IN COMPLEX WITH H-2KB/VSV8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN ), PROTEIN (CD8A OR LYT2 OR LYT-2), ... | | Authors: | Wang, J.H, Reinherz, E.L, Kern, P.S, Chang, H.C. | | Deposit date: | 1998-08-16 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of CD8 coreceptor function revealed by crystallographic analysis of a murine CD8alphaalpha ectodomain fragment in complex with H-2Kb.
Immunity, 9, 1998
|
|
6PAZ
 
 | | OXIDIZED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
3ZG6
 
 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
8DEY
 
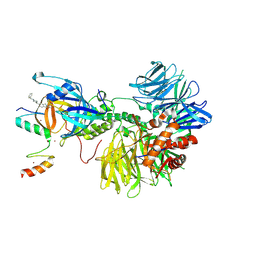 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
7PAZ
 
 | | REDUCED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
1D9K
 
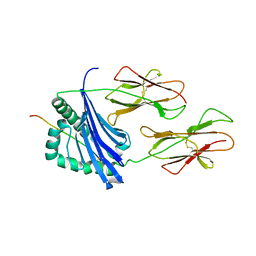 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
5PAZ
 
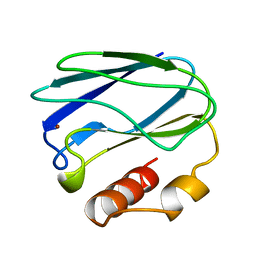 | | REDUCED MUTANT P80A PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
7U8F
 
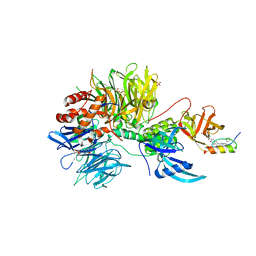 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
6EG3
 
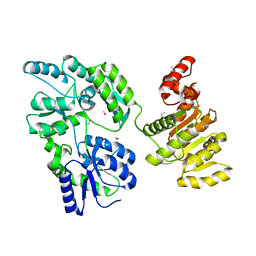 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
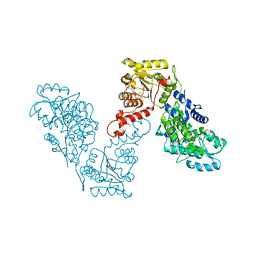 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
1NFD
 
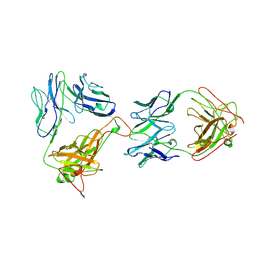 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1WKQ
 
 | |
1Z1J
 
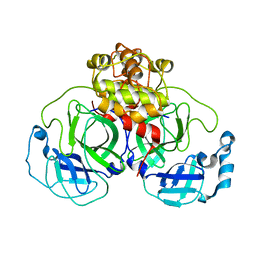 | | Crystal structure of SARS 3CLpro C145A mutant | | Descriptor: | 3C-like proteinase | | Authors: | Hsu, M.F. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
1Z1I
 
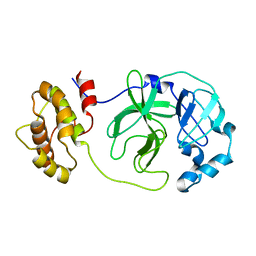 | | Crystal structure of native SARS CLpro | | Descriptor: | 3C-like proteinase | | Authors: | Liang, P.H, Wang, A.H. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
8PAZ
 
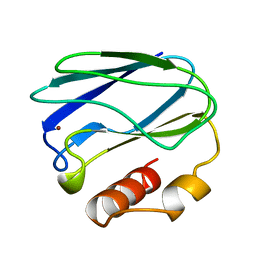 | | OXIDIZED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-24 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
1OSZ
 
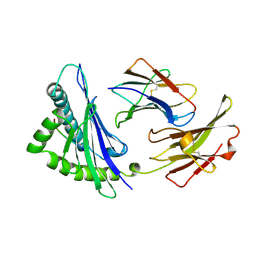 | | MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND AN (L4V) MUTANT OF THE VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2KB HEAVY CHAIN, VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Authors: | Ghendler, Y, Teng, M.-K, Liu, J.-H, Witte, T, Liu, J, Kim, K.S, Kern, P, Chang, H.-C, Wang, J.-H, Reinherz, E.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3FBD
 
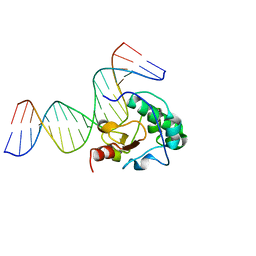 | |
3PAZ
 
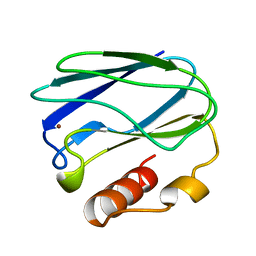 | | REDUCED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
4PAZ
 
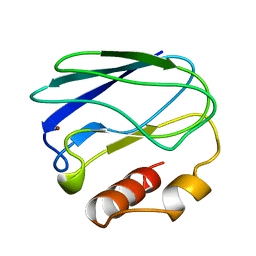 | | OXIDIZED MUTANT P80A PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
