7XQP
 
 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
3JSN
 
 | |
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
7NQ6
 
 | |
8J2W
 
 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
2BV3
 
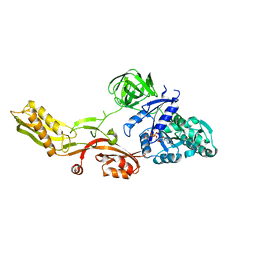 | | Crystal structure of a mutant elongation factor G trapped with a GTP analogue | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Mutant Elongation Factor G Trapped with a GTP Analogue.
FEBS Lett., 579, 2005
|
|
2BM1
 
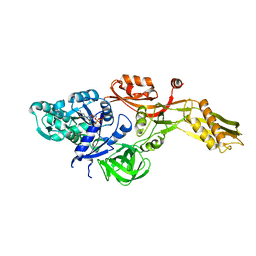 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant G16V | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BM0
 
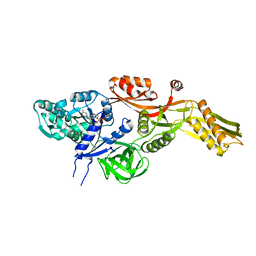 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
3K6U
 
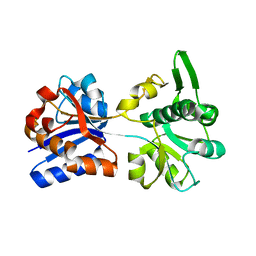 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8J64
 
 | | Crystal structure of Toxoplasma gondii MIC2-M2AP complex | | Descriptor: | MIC2-associated protein, Micronemal protein MIC2 | | Authors: | Zhang, S, Wang, F.F, Zhang, D.J, Song, G.J, Springer, T.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
7BUB
 
 | | Cryo-EM structure of Dengue virus serotype 2 complexed with Fab SIgN-3C at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue virus serotype 2 M protein, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
7BUF
 
 | | Cryo-EM structure of Dengue virus serotype 2 complexed with SIgN-3C IgG | | Descriptor: | Dengue virus serotype 2 E protein, Dengue virus serotype 2 M protein, SIgN-3C IgG heavy chain, ... | | Authors: | Zhang, S, Chew, S.V, Lim, X.N, Ng, T.S, Kostyuchenko, V.A, Lok, S.M. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Cell Rep, 31, 2020
|
|
1SE2
 
 | |
3JSL
 
 | |
3FZO
 
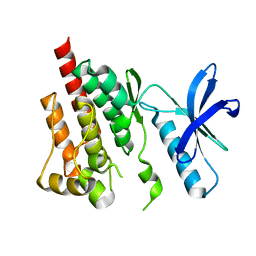 | | Crystal Structure of PYK2-Apo, Proline-rich Tyrosine Kinase | | Descriptor: | Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZR
 
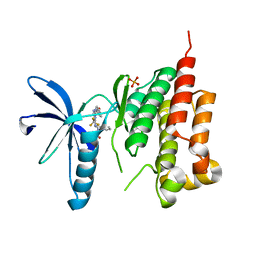 | | Crystal structure of PYK2 complexed with PF-431396 | | Descriptor: | N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZS
 
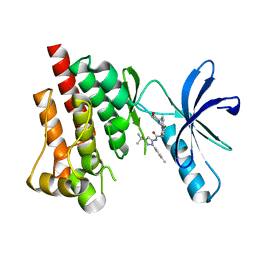 | | Crystal Structure of PYK2 complexed with BIRB796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
4WBC
 
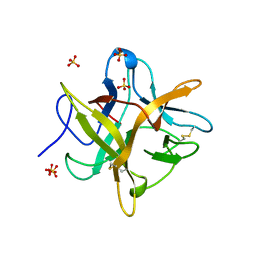 | | 2.13 A STRUCTURE OF A KUNITZ-TYPE WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN | | Descriptor: | PROTEIN (CHYMOTRYPSIN INHIBITOR), SULFATE ION | | Authors: | Ravichandran, S, Sen, U, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Cryocrystallography of a Kunitz-type serine protease inhibitor: the 90 K structure of winged bean chymotrypsin inhibitor (WCI) at 2.13 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3PBO
 
 | | Crystal structure of PBP3 complexed with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBT
 
 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBN
 
 | | Crystal Structure of Apo PBP3 from Pseudomonas aeruginosa | | Descriptor: | Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBS
 
 | | Crystal structure of PBP3 complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBQ
 
 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBR
 
 | | Crystal structure of PBP3 complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XYN
 
 | | The crystal structure of Csm2-Psy3-Shu1-Shu2 complex from budding yeast | | Descriptor: | Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3, Suppressor of HU sensitivity involved in recombination protein 1, ... | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2017-07-09 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the functional role of the Shu complex in homologous recombination.
Nucleic Acids Res., 45, 2017
|
|
