4FSC
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in its apo form | | Descriptor: | Transcriptional activator PlcR protein | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2012-06-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2BZB
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2C0S
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
3U3W
 
 | | Crystal Structure of Bacillus thuringiensis PlcR in complex with the peptide PapR7 and DNA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*AP*AP*AP*TP*AP*TP*TP*GP*CP*AP*TP*AP*G)-3', 5'-D(P*CP*TP*AP*TP*GP*CP*AP*AP*TP*AP*TP*TP*TP*CP*AP*TP*AP*T)-3', C-terminus heptapeptide from PapR protein, ... | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2011-10-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1XE3
 
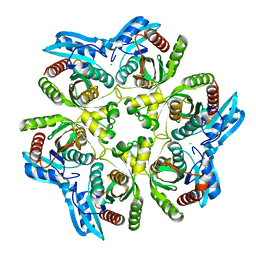 | | Crystal Structure of purine nucleoside phosphorylase DeoD from Bacillus anthracis | | Descriptor: | CHLORIDE ION, purine nucleoside phosphorylase | | Authors: | Grenha, R, Levdikov, V.M, Fogg, M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of purine nucleoside phosphorylase (DeoD) from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
1OF0
 
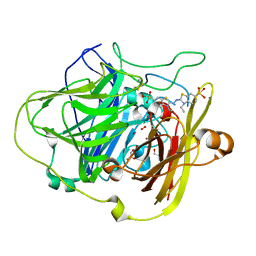 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS COTA AFTER 1H SOAKING WITH ABTS | | Descriptor: | 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (I) ION, CU-O LINKAGE, ... | | Authors: | Enguita, F.J, Marcal, D, Grenha, R, Martins, L.O, Henriques, A.O, Carrondo, M.A. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Characterization of a Bacterial Laccase Reaction Cycle
To be Published
|
|
3ZDW
 
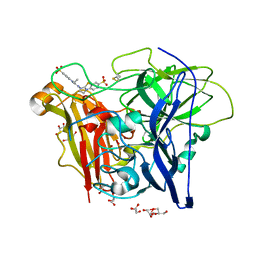 | | Substrate and dioxygen binding to the endospore coat laccase CotA from Bacillus subtilis | | Descriptor: | 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (I) ION, COTA LACCASE, ... | | Authors: | Enguita, F.J, Marcal, D, Grenha, R, Lindley, P.F, Carrondo, M.A. | | Deposit date: | 2012-12-01 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate and Dioxygen Binding to the Endospore Coat Laccase from Bacillus Subtilis.
J.Biol.Chem., 279, 2004
|
|
7OLY
 
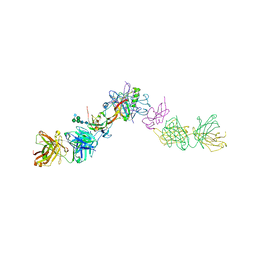 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
6MAC
 
 | | Ternary structure of GDF11 bound to ActRIIB-ECD and Alk5-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 11, ... | | Authors: | Goebel, E.J, Thompson, T.B. | | Deposit date: | 2018-08-27 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7MRZ
 
 | | Structure of GDF11 bound to fused ActRIIB-ECD and Alk4-ECD with Anti-ActRIIB Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B,Activin receptor type-1B, Fab Heavy Chain, ... | | Authors: | Goebel, E.J, Kattamuri, C, Gipson, G.R, Thompson, T.B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
5BJT
 
 | |
1OAE
 
 | | Crystal structure of the reduced form of cytochrome c" from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C", GLYCEROL, HEME C, ... | | Authors: | Enguita, F.J, Grenha, R, Santos, H, Carrondo, M.A. | | Deposit date: | 2003-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
