1QZ8
 
 | | Crystal structure of SARS coronavirus NSP9 | | Descriptor: | SULFATE ION, polyprotein 1ab | | Authors: | Egloff, M.P, Ferron, F, Campanacci, V, Longhi, S, Rancurel, C, Dutartre, H, Snijder, E.J, Gorbalenya, A.E, Cambillau, C, Canard, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1MBM
 
 | | NSP4 proteinase from Equine Arteritis Virus | | Descriptor: | chymotrypsin-like serine protease | | Authors: | Barrette-Ng, I.H, Ng, K.K.-S, Mark, B.L, van Aken, D, Cherney, M.M, Garen, C, Kolodenko, Y, Gorbalenya, A.E, Snijder, E.J, James, M.N.G. | | Deposit date: | 2002-08-03 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Arterivirus nsp4: the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole
J.Biol.Chem., 277, 2002
|
|
1AFV
 
 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3MP2
 
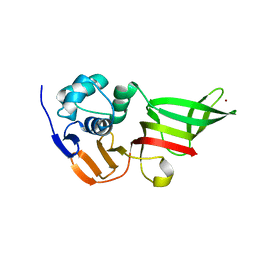 | |
4N0N
 
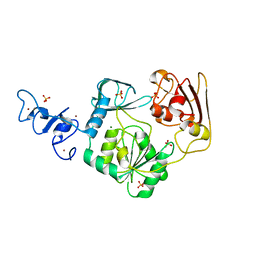 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0O
 
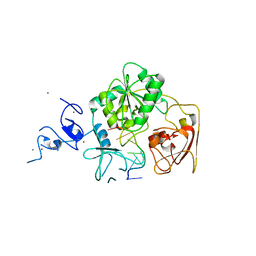 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
5LAC
 
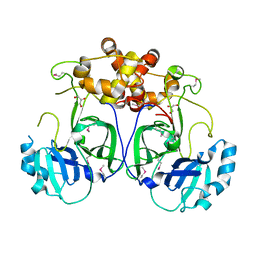 | |
5LAK
 
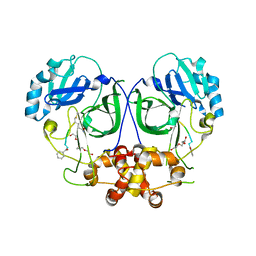 | | Ligand-bound structure of Cavally Virus 3CL Protease | | Descriptor: | 3Cl Protease, BEZ-TYR-TYR-ASN-ECC Peptide inhibitor, PENTAETHYLENE GLYCOL, ... | | Authors: | Kanitz, M, Heine, A, Diederich, W.E. | | Deposit date: | 2016-06-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus.
Virology, 533, 2019
|
|
3Q3Y
 
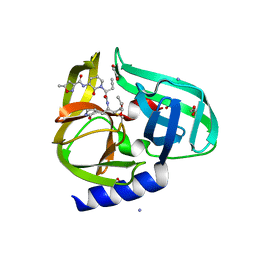 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3Q3X
 
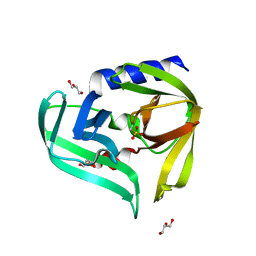 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
5ISN
 
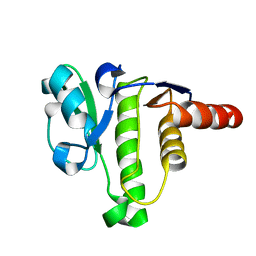 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
3GQE
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus | | Descriptor: | BICINE, Non-structural protein 3 | | Authors: | Jamal, S, Malet, H, Coutard, B, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GQO
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Ferron, F, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GPQ
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with RNA | | Descriptor: | Non-structural protein 3, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GPO
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GPG
 
 | |
3JZT
 
 | | Structure of a cubic crystal form of X (ADRP) domain from FCoV with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2009-09-24 | | Release date: | 2010-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ETI
 
 | |
3EW5
 
 | | Structure of the tetragonal crystal form of X (ADRP) domain from FCoV | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-1-PHOSPHATE, SULFATE ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2008-10-14 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EJG
 
 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3EKE
 
 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3EJF
 
 | |
1LVO
 
 | | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIETHYLENE DIOXIDE, Replicase, ... | | Authors: | Anand, K, Palm, G.J, Mesters, J.R, Siddell, S.G, Ziebuhr, J, Hilgenfeld, R. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain.
EMBO J., 21, 2002
|
|
