2CL8
 
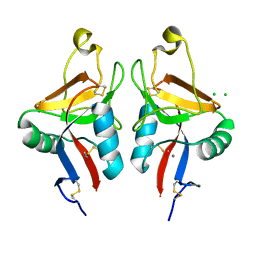 | | Dectin-1 in complex with beta-glucan | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2006-04-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2WJX
 
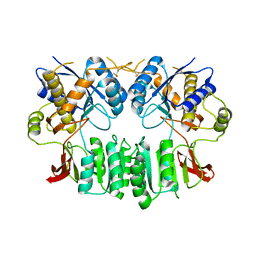 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 4.1 A resolution | | Descriptor: | GLUTAMATE RECEPTOR 2 | | Authors: | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2009-06-01 | | Release date: | 2009-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
2WJW
 
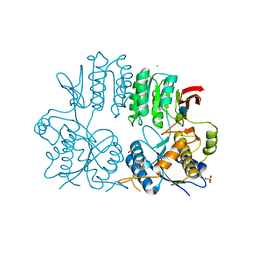 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2009-06-01 | | Release date: | 2009-08-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
2X44
 
 | | Structure of a strand-swapped dimeric form of CTLA-4 | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE PROTEIN 4 | | Authors: | Sonnen, A.F.-P, Yu, C, Evans, E.J, Stuart, D.I, Davis, S.J, Gilbert, R.J.C. | | Deposit date: | 2010-01-28 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Metastability: A Molecular Basis for Immunoglobulin Deposition?
J.Mol.Biol., 399, 2010
|
|
3ZX7
 
 | | Complex of lysenin with phosphocholine | | Descriptor: | LYSENIN, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-09-19 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
3ZXG
 
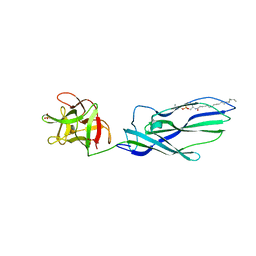 | | lysenin sphingomyelin complex | | Descriptor: | LYSENIN, SULFATE ION, TRIMETHYL-[2-[[(2S,3S)-2-(OCTADECANOYLAMINO)-3-OXIDANYL-BUTOXY]-OXIDANYL-PHOSPHORYL]OXYETHYL]AZANIUM | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-10 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
3ZXD
 
 | | wild-type lysenin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-09 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
4BBK
 
 | | Structural and functional characterisation of the kindlin-1 pleckstrin homology domain | | Descriptor: | FERMITIN FAMILY HOMOLOG 1, GLYCEROL | | Authors: | Yates, L.A, Lumb, C.N, Brahme, N.N, Zalyte, R, Bird, L.E, De Colibus, L, Owens, R.J, Calderwood, D.A, Sansom, M.S.P, Gilbert, R.J.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterisation of the Kindlin-1 Pleckstrin Homology Domain
J.Biol.Chem., 287, 2012
|
|
4C4N
 
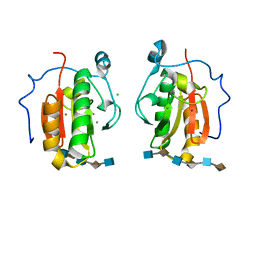 | | Crystal structure of the Sonic Hedgehog-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C4M
 
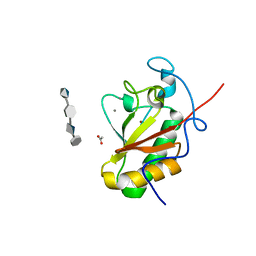 | | Crystal structure of the Sonic Hedgehog-chondroitin-4-sulphate complex | | Descriptor: | ACETATE ION, CALCIUM ION, SONIC HEDGEHOG PROTEIN, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BQB
 
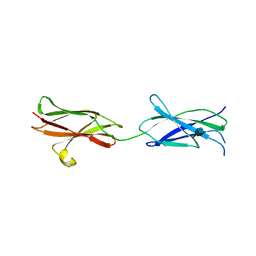 | | Crystal structure of the FN5 and FN6 domains of NEO1, form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQ6
 
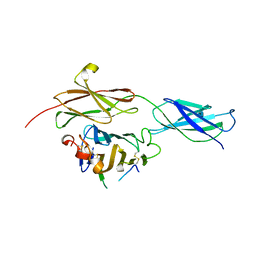 | | Crystal structure of the RGMB-NEO1 complex form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQ9
 
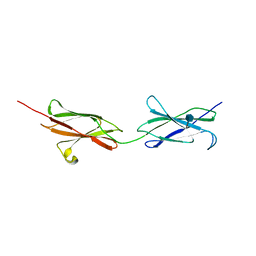 | | Crystal structure of the FN5 and FN6 domains of NEO1, form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQ7
 
 | | Crystal structure of the RGMB-Neo1 complex form 2 | | Descriptor: | NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (6.601 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQ8
 
 | | Crystal structure of the RGMB-NEO1 complex form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BQC
 
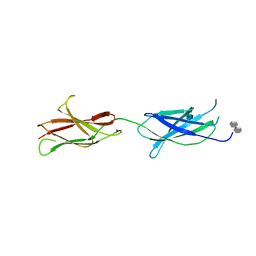 | | Crystal structure of the FN5 and FN6 domains of NEO1 bound to SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, ... | | Authors: | Bell, C.H, Healey, E, vanErp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
8A1D
 
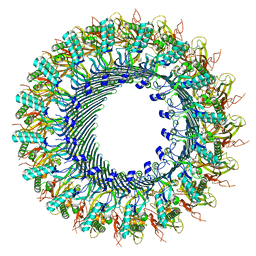 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1S
 
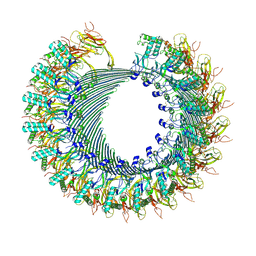 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
4UD4
 
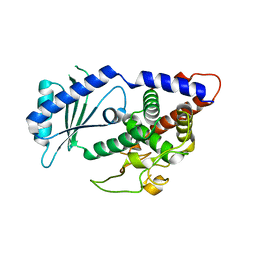 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | GLYCEROL, POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.
Nucleic Acids Res., 43, 2015
|
|
7PLN
 
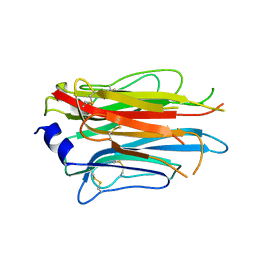 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
5J67
 
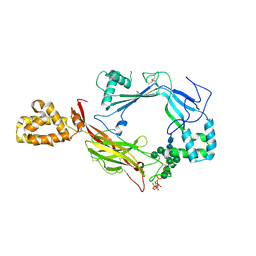 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
5J69
 
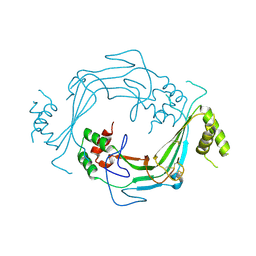 | |
5J68
 
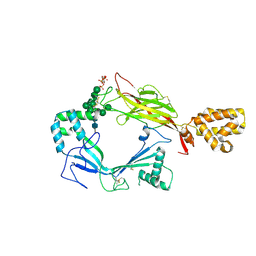 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.221 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
5FN7
 
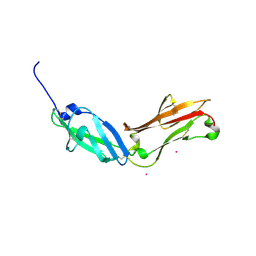 | | Crystal structure of human CD45 extracellular region, domains d1-d2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FMV
 
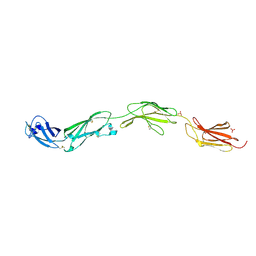 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
