8GCK
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
6NSV
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6XMJ
 
 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | Descriptor: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7M
 
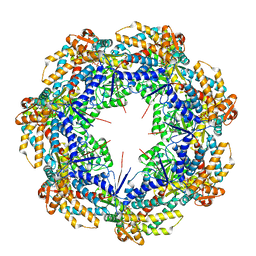 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7N
 
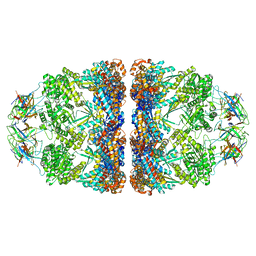 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
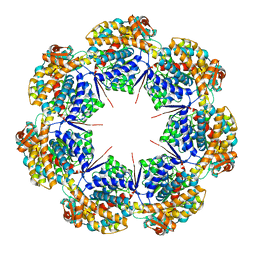 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | Descriptor: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84839141 Å) | | Cite: | Phosphorylation of a Cleaved Tau Proteoform at a Single Residue Inhibits Binding to the E3 Ubiquitin Ligase, CHIP.
Biorxiv, 2023
|
|
8SUV
 
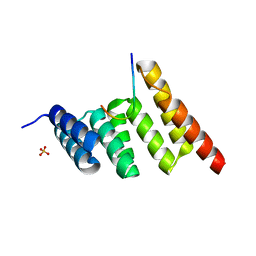 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
2KHO
 
 | |
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7L7J
 
 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7L7I
 
 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
5RSO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000000226 | | Descriptor: | Non-structural protein 3, PARA ACETAMIDO BENZOIC ACID | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RT7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000015442276 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388302 | | Descriptor: | 4-PIPERIDINO-PIPERIDINE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RS9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000007636250 | | Descriptor: | 6,7-dihydro-5H-cyclopenta[d][1,2,4]triazolo[1,5-a]pyrimidin-8-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU6
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001442764 | | Descriptor: | Non-structural protein 3, naphthalene-2-carboximidamide | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
