5V90
 
 | |
1JGN
 
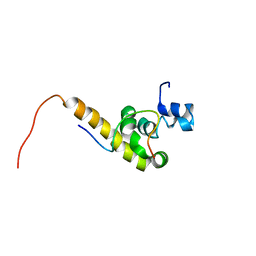 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1JH4
 
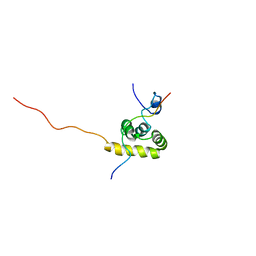 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1DU6
 
 | |
1HO6
 
 | |
1HOQ
 
 | | CHIMERIC RNA/DNA HAIRPIN | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*C)-D(P*TP*TP*CP*GP*GP*TP*CP*C)-3') | | Authors: | Denisov, A.Y, Gehring, K. | | Deposit date: | 2000-12-11 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2'-fluoro-ANA/RNA and DNA/RNA hybrids.
Nucleic Acids Res., 29, 2001
|
|
2ILX
 
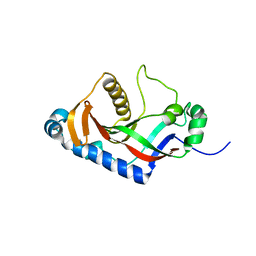 | |
2K18
 
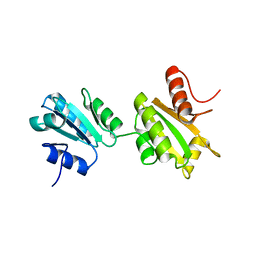 | | Solution structure of bb' domains of human protein disulfide isomerase | | Descriptor: | Protein disulfide-isomerase | | Authors: | Denisov, A.Y, Maattanen, P, Dabrowski, C, Kozlov, G, Thomas, D.Y, Gehring, K. | | Deposit date: | 2008-02-22 | | Release date: | 2008-04-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the bb' domains of human protein disulfide isomerase.
Febs J., 276, 2009
|
|
3KTR
 
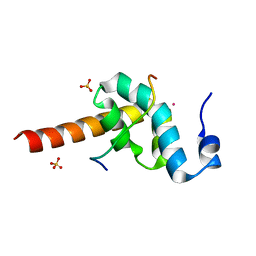 | |
1G9L
 
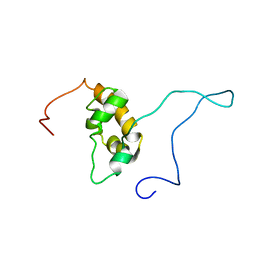 | | SOLUTION STRUCTURE OF THE PABC DOMAIN OF HUMAN POLY(A) BINDING PROTEIN | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1 | | Authors: | Kozlov, G, Trempe, J.-F, Khaleghpour, K, Kahvejian, A, Ekiel, I, Gehring, K. | | Deposit date: | 2000-11-24 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the C-terminal PABC domain of human poly(A)-binding protein.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3KTP
 
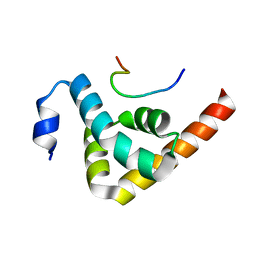 | | Structural basis of GW182 recognition by poly(A)-binding protein | | Descriptor: | Polyadenylate-binding protein 1, Trinucleotide repeat-containing gene 6C protein | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein.
J.Biol.Chem., 285, 2010
|
|
3KUT
 
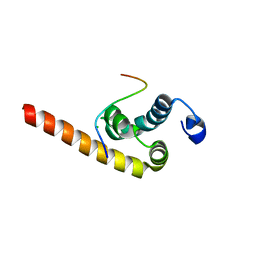 | |
3KUJ
 
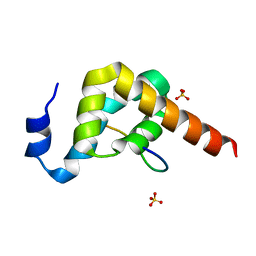 | |
3KUS
 
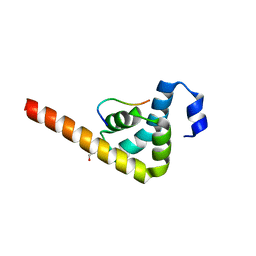 | |
3KUI
 
 | |
3KUR
 
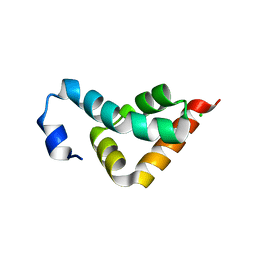 | |
2KBW
 
 | | Solution Structure of human Mcl-1 complexed with human Bid_BH3 peptide | | Descriptor: | BH3-interacting domain death agonist, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, Q, Moldoveanu, T, Sprules, T, Matta-Camacho, E, Mansur-Azzam, N, Gehring, K. | | Deposit date: | 2008-12-09 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Apoptotic regulation by MCL-1 through heterodimerization.
J.Biol.Chem., 285, 2010
|
|
2MJC
 
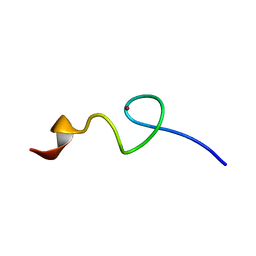 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
3NTW
 
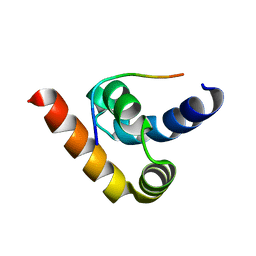 | |
3NY1
 
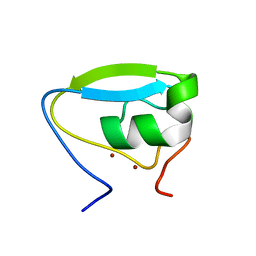 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O0X
 
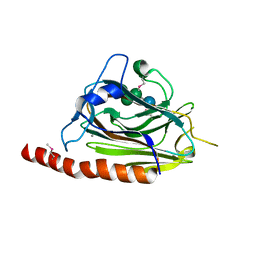 | | Structural basis of carbohydrate recognition by calreticulin | | Descriptor: | CALCIUM ION, Calreticulin, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carbohydrate recognition by calreticulin.
J.Biol.Chem., 285, 2010
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O0W
 
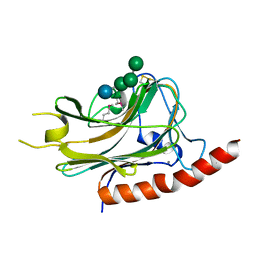 | | Structural basis of carbohydrate recognition by calreticulin | | Descriptor: | CALCIUM ION, Calreticulin, alpha-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of carbohydrate recognition by calreticulin.
J.Biol.Chem., 285, 2010
|
|
3NY3
 
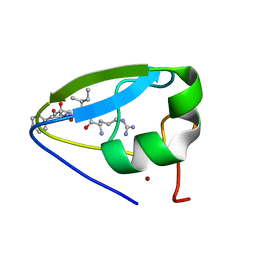 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3O0V
 
 | |
