3KUT
 
 | |
3KUJ
 
 | |
3KUS
 
 | |
3KUI
 
 | |
3KUR
 
 | |
6WUS
 
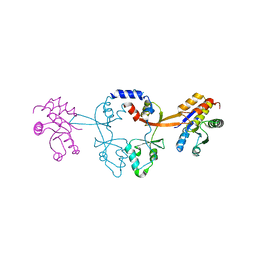 | |
8G91
 
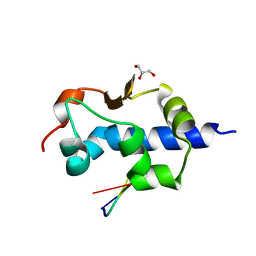 | |
8G90
 
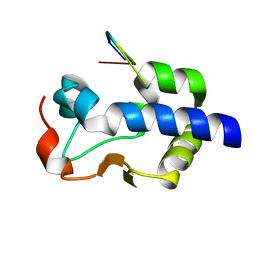 | |
8F6D
 
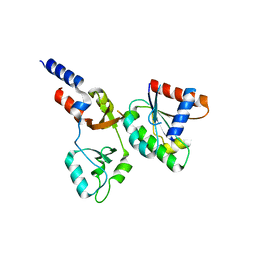 | |
8EY8
 
 | |
8EY6
 
 | |
8EY7
 
 | |
3DF0
 
 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
5K35
 
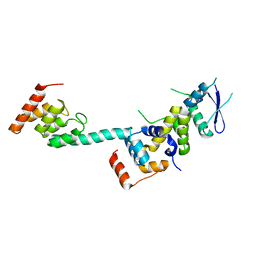 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
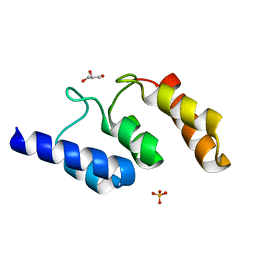 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
4EF0
 
 | |
6DJW
 
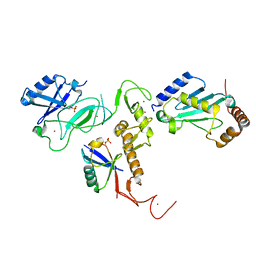 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DFD
 
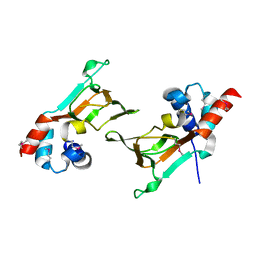 | |
6DJX
 
 | | Crystal Structure of pParkin-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-27 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3BCY
 
 | | Crystal structure of YER067W | | Descriptor: | Protein YER067W | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional study of YER067W, a new protein involved in yeast metabolism control and drug resistance.
Plos One, 5, 2010
|
|
3BXY
 
 | | Crystal structure of tetrahydrodipicolinate N-succinyltransferase from E. coli | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli tetrahydrodipicolinate N-succinyltransferase reveals the role of a conserved C-terminal helix in cooperative substrate binding.
Febs Lett., 582, 2008
|
|
3EC3
 
 | | Crystal structure of the bb fragment of ERp72 | | Descriptor: | Protein disulfide-isomerase A4 | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2008-08-28 | | Release date: | 2009-04-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Noncatalytic Domains and Global Fold of the Protein Disulfide Isomerase ERp72.
Structure, 17, 2009
|
|
5TDA
 
 | |
