5IIS
 
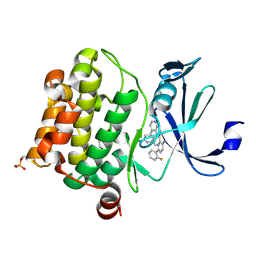 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2A38
 
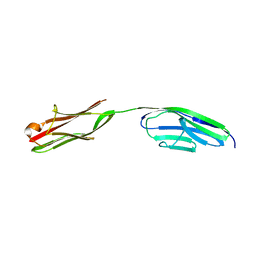 | | Crystal structure of the N-Terminus of titin | | Descriptor: | CADMIUM ION, Titin | | Authors: | Marino, M, Muhle-Goll, C, Svergun, D, Demirel, M, Mayans, O. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin
Structure, 14, 2006
|
|
7Q4T
 
 | |
7Q4S
 
 | |
6CCY
 
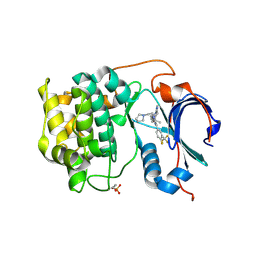 | | Crystal structure of Akt1 in complex with a selective inhibitor | | Descriptor: | (5R)-4-(4-{4-[4-fluoro-3-(trifluoromethyl)phenyl]-1-[2-(pyrrolidin-1-yl)ethyl]-1H-imidazol-2-yl}piperidin-1-yl)-5-methyl-5,8-dihydropyrido[2,3-d]pyrimidin-7(6H)-one, RAC-alpha serine/threonine-protein kinase,PIFtide | | Authors: | Wang, Y, Stout, S. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-02 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of chiral dihydropyridopyrimidinones as potent, selective and orally bioavailable inhibitors of AKT.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6HRG
 
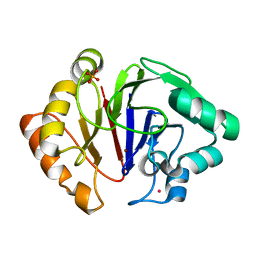 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
7Z6B
 
 | |
8A2C
 
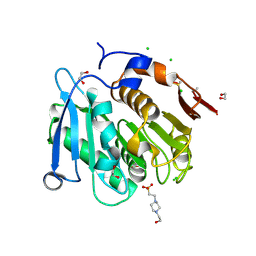 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 hydrolyses polyethylene terephthalate (PET) and is well conserved in the GC-rich gram-positives Amycolatopsis and Streptomyces
To Be Published
|
|
2X8P
 
 | | Crystal Structure of CbpF in Complex with Atropine by Co- Crystallization | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CHOLINE ION, CHOLINE-BINDING PROTEIN F, ... | | Authors: | Silva-Martin, N, Hermoso, J.A. | | Deposit date: | 2010-03-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of Cbpf Complexed with Atropine and Ipratropium Reveal Clues for the Design of Novel Antimicrobials Against Streptococcus Pneumoniae.
Biochim.Biophys.Acta, 1840, 2013
|
|
2X8M
 
 | |
5LQQ
 
 | | Structure of Autotaxin (ENPP2) with LM350 | | Descriptor: | 3-(6-chloranyl-2-methyl-1-phenyl-indol-3-yl)sulfanylbenzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.J, Heidebrecht, T, Castelmur, E, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-08-17 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Activity Relationships of Small Molecule Autotaxin Inhibitors with a Discrete Binding Mode.
J. Med. Chem., 60, 2017
|
|
2JTO
 
 | | Solution Structure of Tick Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase inhibitor | | Authors: | Pantoja-Uceda, D, Blanco, F.J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-07-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR structure and dynamics of the two-domain tick carboxypeptidase inhibitor reveal flexibility in its free form and stiffness upon binding to human carboxypeptidase B.
Biochemistry, 47, 2008
|
|
6EXZ
 
 | | Crystal structure of Mex67 C-term | | Descriptor: | FORMIC ACID, mRNA export factor MEX67 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mip6 binds directly to the Mex67 UBA domain to maintain low levels of Msn2/4 stress-dependent mRNAs.
Embo Rep., 2019
|
|
