1EJB
 
 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
3DDY
 
 | | Structure of lumazine protein, an optical transponder of luminescent bacteria | | Descriptor: | Lumazine protein, RIBOFLAVIN | | Authors: | Chatwell, L, Illarionova, V, Illarionov, B, Skerra, A, Bacher, A, Fischer, M. | | Deposit date: | 2008-06-07 | | Release date: | 2008-07-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of lumazine protein, an optical transponder of luminescent bacteria.
J.Mol.Biol., 382, 2008
|
|
5UG2
 
 | | CcP gateless cavity | | Descriptor: | 6-fluoro-2-methylimidazo[1,2-a]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Stein, R.M, Fischer, M, Shoichet, B.K. | | Deposit date: | 2017-01-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WTQ
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTN
 
 | | Human JAK2 JH1 domain in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTP
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 3 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, tert-butyl 4-[(4-{1-[3-(cyanomethyl)-1-(ethylsulfonyl)azetidin-3-yl]-1H-pyrazol-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino]benzoate | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTO
 
 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
1X9R
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the oxidised form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
1X9U
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the reduced form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
1W29
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1PVW
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from M. jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Steinbacher, S, Schiffmann, S, Richter, G, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2003-06-29 | | Release date: | 2003-11-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of 3,4-Dihydroxy-2-butanone 4-Phosphate Synthase from Methanococcus jannaschii in
Complex with Divalent Metal Ions and the Substrate Ribulose 5-Phosphate: IMPLICATIONS FOR THE
CATALYTIC MECHANISM
J.Biol.Chem., 278, 2003
|
|
1PVY
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from M. jannaschii in complex with ribulose 5-phosphate | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Richter, G, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2003-06-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of 3,4-Dihydroxy-2-butanone 4-Phosphate Synthase from Methanococcus jannaschii in
Complex with Divalent Metal Ions and the Substrate Ribulose 5-Phosphate: IMPLICATIONS FOR THE
CATALYTIC MECHANISM
J.Biol.Chem., 278, 2003
|
|
1W19
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1SEP
 
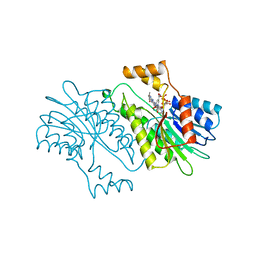 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1ITZ
 
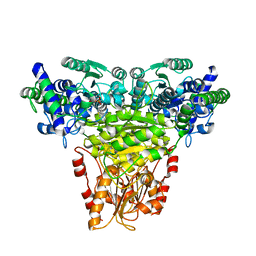 | | Maize Transketolase in complex with TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Gerhardt, S, Echt, S, Bader, G, Freigang, J, Busch, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-15 | | Release date: | 2003-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and properties of an engineered transketolase from maize
PLANT PHYSIOL., 132, 2003
|
|
1JG5
 
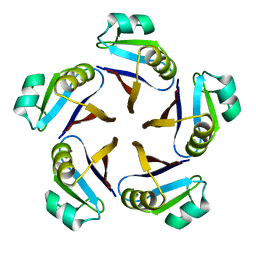 | | CRYSTAL STRUCTURE OF RAT GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, GFRP | | Descriptor: | GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, POTASSIUM ION | | Authors: | Bader, G, Schiffmann, S, Herrmann, A, Fischer, M, Gutlich, M, Auerbach, G, Ploom, T, Bacher, A, Huber, R, Lemm, T. | | Deposit date: | 2001-06-23 | | Release date: | 2001-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat GTP cyclohydrolase I feedback regulatory protein, GFRP.
J.Mol.Biol., 312, 2001
|
|
1KZ6
 
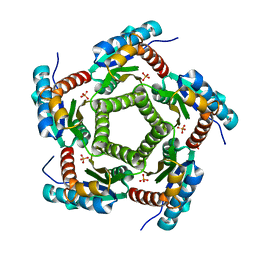 | | Mutant enzyme W63Y/L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KYY
 
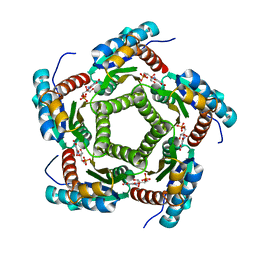 | | Lumazine Synthase from S.pombe bound to nitropyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KZ1
 
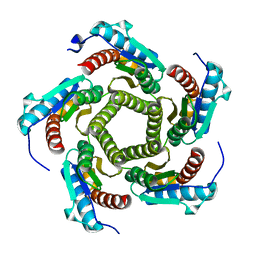 | | Mutant enzyme W27G Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KZ9
 
 | | Mutant Enzyme L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KYX
 
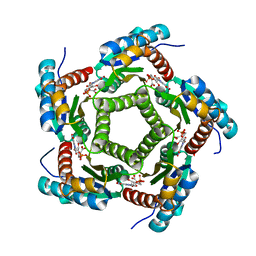 | | Lumazine Synthase from S.pombe bound to carboxyethyllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KZ4
 
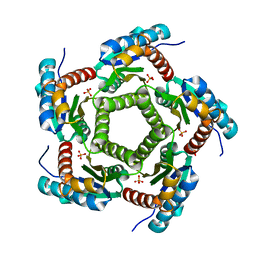 | | Mutant enzyme W63Y Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KYV
 
 | | Lumazine Synthase from S.pombe bound to riboflavin | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KZL
 
 | | Riboflavin Synthase from S.pombe bound to Carboxyethyllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, MERCURY (II) ION, Riboflavin Synthase | | Authors: | Gerhardt, S, Schott, A.K, Kairies, N, Cushman, M, Illarionov, B, Eisenreich, W, Bacher, A, Huber, R, Steinbacher, S, Fischer, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on the Reaction Mechanism of Riboflavin Synthase; X-Ray Crystal Structure of a Complex with 6-Carboxyethyl-7-Oxo-8-Ribityllumazine
STRUCTURE, 10, 2002
|
|
4NAL
 
 | | Arabidopsis thaliana IspD in complex with tribromodichloro-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
